DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins
- PMID: 27466170
- PMCID: PMC4964356
- DOI: 10.1038/srep30620
DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins
Abstract
Microalgae are versatile organisms capable of converting CO2, H2O, and sunlight into fuel and chemicals for domestic and industrial consumption. Thus, genetic modifications of microalgae for enhancing photosynthetic productivity, and biomass and bio-products generation are crucial for both academic and industrial applications. However, targeted mutagenesis in microalgae with CRISPR-Cas9 is limited. Here we report, a one-step transformation of Chlamydomonas reinhardtii by the DNA-free CRISPR-Cas9 method rather than plasmids that encode Cas9 and guide RNAs. Outcome was the sequential CpFTSY and ZEP two-gene knockout and the generation of a strain constitutively producing zeaxanthin and showing improved photosynthetic productivity.
Conflict of interest statement
K.B., D.H.K., E.J. and S.B. are co-inventors on a patent application covering the genome editing method described in this manuscript.
Figures
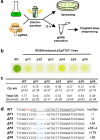
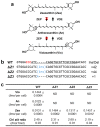

References
-
- Wijffels R. H. & Barbosa M. J. An outlook on microalgal biofuels. Science 329, 796–799 (2010). - PubMed
-
- Koller M., Muhr A. & Braunegg G. Microalgae as versatile cellular factories for valued products. Algal Res. 6, 52–63 (2014).
-
- Sizova I., Greiner A., Awasthi M., Kateriya S. & Hegemann P. Nuclear gene targeting in Chlamydomonas using engineered zinc‐finger nucleases. Plant J. 73, 873–882 (2013). - PubMed
-
- Daboussi F. et al.. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun. 5, (2014). - PubMed
-
- Gao H. et al.. TALE activation of endogenous genes in Chlamydomonas reinhardtii. Algal Res. 5, 52–60 (2014).

