The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers
- PMID: 27466363
- PMCID: PMC5034058
- DOI: 10.1074/jbc.M116.739136
The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers
Abstract
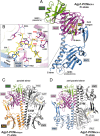
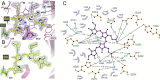
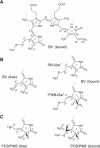
Agp1 is a canonical biliverdin-binding bacteriophytochrome from the soil bacterium Agrobacterium fabrum that acts as a light-regulated histidine kinase. Crystal structures of the photosensory core modules (PCMs) of homologous phytochromes have provided a consistent picture of the structural changes that these proteins undergo during photoconversion between the parent red light-absorbing state (Pr) and the far-red light-absorbing state (Pfr). These changes include secondary structure rearrangements in the so-called tongue of the phytochrome-specific (PHY) domain and structural rearrangements within the long α-helix that connects the cGMP-specific phosphodiesterase, adenylyl cyclase, and FhlA (GAF) and the PHY domains. We present the crystal structures of the PCM of Agp1 at 2.70 Å resolution and of a surface-engineered mutant of this PCM at 1.85 Å resolution in the dark-adapted Pr states. Whereas in the mutant structure the dimer subunits are in anti-parallel orientation, the wild-type structure contains parallel subunits. The relative orientations between the PAS-GAF bidomain and the PHY domain are different in the two structures, due to movement involving two hinge regions in the GAF-PHY connecting α-helix and the tongue, indicating pronounced structural flexibility that may give rise to a dynamic Pr state. The resolution of the mutant structure enabled us to detect a sterically strained conformation of the chromophore at ring A that we attribute to the tight interaction with Pro-461 of the conserved PRXSF motif in the tongue. Based on this observation and on data from mutants where residues in the tongue region were replaced by alanine, we discuss the crucial roles of those residues in Pr-to-Pfr photoconversion.
Keywords: Agrobacterium; conformational change; crystal structure; histidine kinase; photoreceptor; phytochrome; quaternary structure; signal transduction; tertiary structure.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

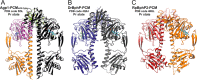



References
-
- Auldridge M. E., and Forest K. T. (2011) Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 - PubMed
-
- Ulijasz A. T., and Vierstra R. D. (2011) Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr. Opin. Plant Biol. 14, 498–506 - PubMed
-
- Vierstra R. D., and Zhang J. (2011) Phytochrome signaling: solving the Gordian knot with microbial relatives. Trends Plant Sci. 16, 417–426 - PubMed
-
- Duanmu D., Bachy C., Sudek S., Wong C. H., Jiménez V., Rockwell N. C., Martin S. S., Ngan C. Y., Reistetter E. N., van Baren M. J., Price D. C., Wei C. L., Reyes-Prieto A., Lagarias J. C., and Worden A. Z. (2014) Marine algae and land plants share conserved phytochrome signaling systems. Proc. Natl. Acad. Sci. U.S.A. 111, 15827–15832 - PMC - PubMed
-
- Bai Y., Rottwinkel G., Feng J., Liu Y., and Lamparter T. (2016) Bacteriophytochromes control conjugation in Agrobacterium fabrum. J. Photochem. Photobiol. B 161, 192–199 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

