Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion
- PMID: 27466418
- PMCID: PMC5021412
- DOI: 10.1128/JVI.01429-16
Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion
Abstract
The highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) cause significant morbidity and morality. There is currently no approved therapeutic for highly pathogenic coronaviruses, even as MERS-CoV is spreading throughout the Middle East. We previously screened a library of FDA-approved drugs for inhibitors of coronavirus replication in which we identified Abelson (Abl) kinase inhibitors, including the anticancer drug imatinib, as inhibitors of both SARS-CoV and MERS-CoV in vitro Here we show that the anti-CoV activity of imatinib occurs at the early stages of infection, after internalization and endosomal trafficking, by inhibiting fusion of the virions at the endosomal membrane. We specifically identified the imatinib target, Abelson tyrosine-protein kinase 2 (Abl2), as required for efficient SARS-CoV and MERS-CoV replication in vitro These data demonstrate that specific approved drugs can be characterized in vitro for their anticoronavirus activity and used to identify host proteins required for coronavirus replication. This type of study is an important step in the repurposing of approved drugs for treatment of emerging coronaviruses.
Importance: Both SARS-CoV and MERS-CoV are zoonotic infections, with bats as the primary source. The 2003 SARS-CoV outbreak began in Guangdong Province in China and spread to humans via civet cats and raccoon dogs in the wet markets before spreading to 37 countries. The virus caused 8,096 confirmed cases of SARS and 774 deaths (a case fatality rate of ∼10%). The MERS-CoV outbreak began in Saudi Arabia and has spread to 27 countries. MERS-CoV is believed to have emerged from bats and passed into humans via camels. The ongoing outbreak of MERS-CoV has resulted in 1,791 cases of MERS and 640 deaths (a case fatality rate of 36%). The emergence of SARS-CoV and MERS-CoV provides evidence that coronaviruses are currently spreading from zoonotic sources and can be highly pathogenic, causing serious morbidity and mortality in humans. Treatment of SARS-CoV and MERS-CoV infection is limited to providing supportive therapy consistent with any serious lung disease, as no specific drugs have been approved as therapeutics. Highly pathogenic coronaviruses are rare and appear to emerge and disappear within just a few years. Currently, MERS-CoV is still spreading, as new infections continue to be reported. The outbreaks of SARS-CoV and MERS-CoV and the continuing diagnosis of new MERS cases highlight the need for finding therapeutics for these diseases and potential future coronavirus outbreaks. Screening FDA-approved drugs streamlines the pipeline for this process, as these drugs have already been tested for safety in humans.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
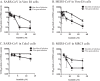


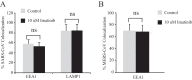
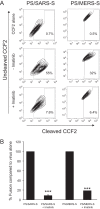
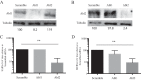
References
-
- Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. doi: 10.1371/journal.ppat.1004502. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

