HIV Maintains an Evolving and Dispersed Population in Multiple Tissues during Suppressive Combined Antiretroviral Therapy in Individuals with Cancer
- PMID: 27466425
- PMCID: PMC5044847
- DOI: 10.1128/JVI.00684-16
HIV Maintains an Evolving and Dispersed Population in Multiple Tissues during Suppressive Combined Antiretroviral Therapy in Individuals with Cancer
Abstract
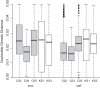
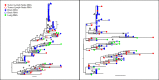
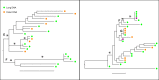
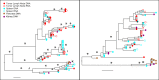
While combined antiretroviral therapy (cART) can result in undetectable plasma viral loads, it does not eradicate HIV infection. Furthermore, HIV-infected individuals while on cART remain at an increased risk of developing serious comorbidities, such as cancer, neurological disease, and atherosclerosis, suggesting that during cART, tissue-based HIV may contribute to such pathologies. We obtained DNA and RNA env, nef, and pol sequences using single-genome sequencing from postmortem tissues of three HIV(+) cART-treated (cART(+)) individuals with undetectable viral load and metastatic cancer at death and performed time-scaled Bayesian evolutionary analyses. We used a sensitive in situ hybridization technique to visualize HIV gag-pol mRNA transcripts in cerebellum and lymph node tissues from one patient. Tissue-associated virus evolved at similar rates in cART(+) and cART-naive (cART(-)) patients. Phylogenetic trees were characterized by two distinct features: (i) branching patterns consistent with constant viral evolution and dispersal among tissues and (ii) very recently derived clades containing both DNA and RNA sequences from multiple tissues. Rapid expansion of virus near death corresponded to wide-spread metastasis. HIV RNA(+) cells clustered in cerebellum tissue but were dispersed in lymph node tissue, mirroring the evolutionary patterns observed for that patient. Activated, infiltrating macrophages were associated with HIV RNA. Our data provide evidence that tissues serve as a sanctuary for wild-type HIV during cART and suggest the importance of macrophages as an alternative reservoir and mechanism of virus spread.
Importance: Combined antiretroviral therapy (cART) reduces plasma HIV to undetectable levels; however, removal of cART results in plasma HIV rebound, thus highlighting its inability to entirely rid the body of infection. Additionally, HIV-infected individuals on cART remain at high risk of serious diseases, which suggests a contribution from residual HIV. In this study, we isolated and sequenced HIV from postmortem tissues from three HIV(+) cART(+) individuals who died with metastatic cancer and had no detectable plasma viral load. Using high-resolution evolutionary analyses, we found that tissue-based HIV continues to replicate, evolve, and migrate among tissues during cART. Furthermore, cancer onset and metastasis coincided with increased HIV expansion, suggesting a linked mechanism. HIV-expressing cells were associated with tissue macrophages, a target of HIV infection. Our results suggest the importance of tissues, and macrophages in particular, as a target for novel anti-HIV therapies.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi:10.1038/nm.1972. - DOI - PMC - PubMed
-
- Chun T, Carruth L, Finzi D, Shen X, DiGiuseppe J, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T, Kuo Y, Brookmeyer R, Zeiger M, Barditch-Crovo P, Siliciano R. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi:10.1038/387183a0. - DOI - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi:10.1038/8394. - DOI - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth L, Buck C, Chaisson R, Quinn T, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D, Richman D, Siliciano R. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

