Dynamic phosphorylation of RelA on Ser42 and Ser45 in response to TNFα stimulation regulates DNA binding and transcription
- PMID: 27466442
- PMCID: PMC4967822
- DOI: 10.1098/rsob.160055
Dynamic phosphorylation of RelA on Ser42 and Ser45 in response to TNFα stimulation regulates DNA binding and transcription
Abstract
The NF-κB signalling module controls transcription through a network of protein kinases such as the IKKs, as well as inhibitory proteins (IκBs) and transcription factors including RelA/p65. Phosphorylation of the NF-κB subunits is critical for dictating system dynamics. Using both non-targeted discovery and quantitative selected reaction monitoring-targeted proteomics, we show that the cytokine TNFα induces dynamic multisite phosphorylation of RelA at a number of previously unidentified residues. Putative roles for many of these phosphorylation sites on RelA were predicted by modelling of various crystal structures. Stoichiometry of phosphorylation determination of Ser45 and Ser42 revealed preferential early phosphorylation of Ser45 in response to TNFα. Quantitative analyses subsequently confirmed differential roles for pSer42 and pSer45 in promoter-specific DNA binding and a role for both of these phosphosites in regulating transcription from the IL-6 promoter. These temporal dynamics suggest that RelA-mediated transcription is likely to be controlled by functionally distinct NF-κB proteoforms carrying different combinations of modifications, rather than a simple 'one modification, one effect' system.
Keywords: NF-κB; RelA; phosphorylation; proteomics; quantification; transcription.
© 2016 The Authors.
Figures



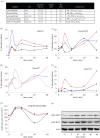
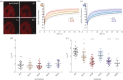
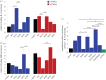
References
-
- Lawrence T. 2009. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1, a001651 (doi:10.1101/cshperspect.a001651) - DOI - PMC - PubMed
-
- Sun SC, Chang JH, Jin J. 2013. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 34, 282–289. (doi:10.1016/j.it.2013.01.004) - DOI - PMC - PubMed
-
- DiDonato JA, Mercurio F, Karin M. 2012. NF-κB and the link between inflammation and cancer. Immunol. Rev. 246, 379–400. (doi:10.1111/j.1600-065X.2012.01099.x) - DOI - PubMed
-
- Neumann M, Naumann M. 2007. Beyond IκBs: alternative regulation of NF-κB activity. FASEB J. 21, 2642–2654. (doi:10.1096/fj.06-7615rev) - DOI - PubMed
-
- Perkins ND. 2006. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene 25, 6717–6730. (doi:10.1038/sj.onc.1209937) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/K003097/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L009501/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K001949/1/MRC_/Medical Research Council/United Kingdom
- BB/F005938/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G009058/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F00561X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT0805090MA/WT_/Wellcome Trust/United Kingdom
- BB/E024181/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U105960399/MRC_/Medical Research Council/United Kingdom
- BB/F005938/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
