Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus
- PMID: 27467578
- PMCID: PMC5026613
- DOI: 10.1016/j.virol.2016.07.016
Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus
Abstract
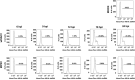
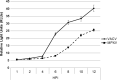
Monkeypox virus (MPXV) infection fails to activate the host anti-viral protein, PKR, despite lacking a full-length homologue of the vaccinia virus (VACV) PKR inhibitor, E3. Since PKR can be activated by dsRNA produced during a viral infection, we have analyzed the accumulation of dsRNA in MPXV-infected cells. MPXV infection led to less accumulation of dsRNA than VACV infection. Because in VACV infections accumulation of abnormally low amounts of dsRNA is associated with mutations that lead to resistance to the anti-poxvirus drug isatin beta-thiosemicarbazone (IBT), we investigated the effects of treatment of MPXV-infected cells with IBT. MPXV infection was eight-fold more resistant to IBT than wild-type vaccinia virus (wtVACV). These results demonstrate that MPXV infection leads to the accumulation of less dsRNA than wtVACV, which in turn likely leads to a decreased capacity for activation of the dsRNA-dependent host enzyme, PKR.
Keywords: E3 gene; F3 gene; IBT resistance; Innate immune evasion; Monkeypox virus; Poxvirus pathogenesis; Poxvirus transcription; Vaccinia virus; Virulence; dsRNA.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





References
-
- Bayliss C.D., Condit R.C. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology. 1993;194:254–262. - PubMed
-
- Brandt T., Heck M.C., Vijaysri S., Jentarra G.M., Cameron J.M., Jacobs B.L. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology. 2005;333:263–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

