Temporal Shift of Circadian-Mediated Gene Expression and Carbon Fixation Contributes to Biomass Heterosis in Maize Hybrids
- PMID: 27467757
- PMCID: PMC4965137
- DOI: 10.1371/journal.pgen.1006197
Temporal Shift of Circadian-Mediated Gene Expression and Carbon Fixation Contributes to Biomass Heterosis in Maize Hybrids
Abstract
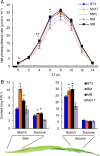
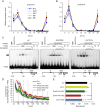
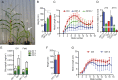
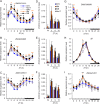
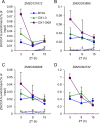
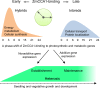
Heterosis has been widely used in agriculture, but the molecular mechanism for this remains largely elusive. In Arabidopsis hybrids and allopolyploids, increased photosynthetic and metabolic activities are linked to altered expression of circadian clock regulators, including CIRCADIAN CLOCK ASSOCIATED1 (CCA1). It is unknown whether a similar mechanism mediates heterosis in maize hybrids. Here we report that higher levels of carbon fixation and starch accumulation in the maize hybrids are associated with altered temporal gene expression. Two maize CCA1 homologs, ZmCCA1a and ZmCCA1b, are diurnally up-regulated in the hybrids. Expressing ZmCCA1 complements the cca1 mutant phenotype in Arabidopsis, and overexpressing ZmCCA1b disrupts circadian rhythms and biomass heterosis. Furthermore, overexpressing ZmCCA1b in maize reduced chlorophyll content and plant height. Reduced height stems from reduced node elongation but not total node number in both greenhouse and field conditions. Phenotypes are less severe in the field than in the greenhouse, suggesting that enhanced light and/or metabolic activities in the field can compensate for altered circadian regulation in growth vigor. Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis reveals a temporal shift of ZmCCA1-binding targets to the early morning in the hybrids, suggesting that activation of morning-phased genes in the hybrids promotes photosynthesis and growth vigor. This temporal shift of ZmCCA1-binding targets correlated with nonadditive and additive gene expression in early and late stages of seedling development. These results could guide breeding better hybrid crops to meet the growing demand in food and bioenergy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The regulatory network mediated by circadian clock genes is related to heterosis in rice.J Integr Plant Biol. 2015 Mar;57(3):300-12. doi: 10.1111/jipb.12240. Epub 2014 Sep 25. J Integr Plant Biol. 2015. PMID: 25040350
-
A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs.Nat Commun. 2021 Apr 19;12(1):2317. doi: 10.1038/s41467-021-22268-z. Nat Commun. 2021. PMID: 33875651 Free PMC article.
-
Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis.Proc Natl Acad Sci U S A. 2018 May 22;115(21):5606-5611. doi: 10.1073/pnas.1722068115. Epub 2018 May 7. Proc Natl Acad Sci U S A. 2018. PMID: 29735680 Free PMC article.
-
Epigenetic Changes in Hybrids.Plant Physiol. 2015 Aug;168(4):1197-205. doi: 10.1104/pp.15.00231. Epub 2015 May 22. Plant Physiol. 2015. PMID: 26002907 Free PMC article. Review.
-
Unlocking allelic variation in circadian clock genes to develop environmentally robust and productive crops.Planta. 2024 Feb 22;259(4):72. doi: 10.1007/s00425-023-04324-8. Planta. 2024. PMID: 38386103 Free PMC article. Review.
Cited by
-
The overexpression of the switchgrass (Panicum virgatum L.) genes PvTOC1-N or PvLHY-K affects circadian rhythm and hormone metabolism in transgenic Arabidopsis seedlings.Biotechnol Biofuels Bioprod. 2024 Oct 3;17(1):126. doi: 10.1186/s13068-024-02574-6. Biotechnol Biofuels Bioprod. 2024. PMID: 39363306 Free PMC article.
-
Shade signals alter the expression of circadian clock genes in newly-formed bioenergy sorghum internodes.Plant Direct. 2020 Jun 25;4(6):e00235. doi: 10.1002/pld3.235. eCollection 2020 Jun. Plant Direct. 2020. PMID: 32607464 Free PMC article.
-
Alternative Splicing of Circadian Clock Genes Correlates With Temperature in Field-Grown Sugarcane.Front Plant Sci. 2019 Dec 23;10:1614. doi: 10.3389/fpls.2019.01614. eCollection 2019. Front Plant Sci. 2019. PMID: 31921258 Free PMC article.
-
Expression Patterns Divergence of Reciprocal F1 Hybrids Between Gossypium hirsutum and Gossypium barbadense Reveals Overdominance Mediating Interspecific Biomass Heterosis.Front Plant Sci. 2022 Jul 1;13:892805. doi: 10.3389/fpls.2022.892805. eCollection 2022. Front Plant Sci. 2022. PMID: 35845678 Free PMC article.
-
Plant height heterosis is quantitatively associated with expression levels of plastid ribosomal proteins.Proc Natl Acad Sci U S A. 2021 Nov 23;118(47):e2109332118. doi: 10.1073/pnas.2109332118. Proc Natl Acad Sci U S A. 2021. PMID: 34782463 Free PMC article.
References
-
- Duvick DN. Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet. 2001;2(1):69–74. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

