Biosynthesis and Metabolic Fate of Phenylalanine in Conifers
- PMID: 27468292
- PMCID: PMC4942462
- DOI: 10.3389/fpls.2016.01030
Biosynthesis and Metabolic Fate of Phenylalanine in Conifers
Abstract
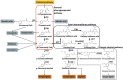
The amino acid phenylalanine (Phe) is a critical metabolic node that plays an essential role in the interconnection between primary and secondary metabolism in plants. Phe is used as a protein building block but it is also as a precursor for numerous plant compounds that are crucial for plant reproduction, growth, development, and defense against different types of stresses. The metabolism of Phe plays a central role in the channeling of carbon from photosynthesis to the biosynthesis of phenylpropanoids. The study of this metabolic pathway is particularly relevant in trees, which divert large amounts of carbon into the biosynthesis of Phe-derived compounds, particularly lignin, an important constituent of wood. The trunks of trees are metabolic sinks that consume a considerable percentage of carbon and energy from photosynthesis, and carbon is finally immobilized in wood. This paper reviews recent advances in the biosynthesis and metabolic utilization of Phe in conifer trees. Two alternative routes have been identified: the ancient phenylpyruvate pathway that is present in microorganisms, and the arogenate pathway that possibly evolved later during plant evolution. Additionally, an efficient nitrogen recycling mechanism is required to maintain sustained growth during xylem formation. The relevance of phenylalanine metabolic pathways in wood formation, the biotic interactions, and ultraviolet protection is discussed. The genetic manipulation and transcriptional regulation of the pathways are also outlined.
Keywords: aromatic amino acids; gene regulatory networks; nitrogen recycling; phenylpropanoids; trees.
Figures


References
-
- Beckman C. H. (2000). Phenolic-storing cells: keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 57 101–110. 10.1006/pmpp.2000.0287 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

