FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis
- PMID: 27469166
- PMCID: PMC4995929
- DOI: 10.1073/pnas.1602960113
FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis
Abstract
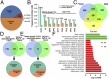
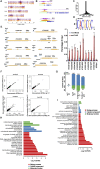
Plant meristems are responsible for the generation of all plant tissues and organs. Here we show that the transcription factor (TF) FAR-RED ELONGATED HYPOCOTYL3 (FHY3) plays an important role in both floral meristem (FM) determinacy and shoot apical meristem maintenance in Arabidopsis, in addition to its well-known multifaceted roles in plant growth and development during the vegetative stage. Through genetic analyses, we show that WUSCHEL (WUS) and CLAVATA3 (CLV3), two central players in the establishment and maintenance of meristems, are epistatic to FHY3 Using genome-wide ChIP-seq and RNA-seq data, we identify hundreds of FHY3 target genes in flowers and find that FHY3 mainly acts as a transcriptional repressor in flower development, in contrast to its transcriptional activator role in seedlings. Binding motif-enrichment analyses indicate that FHY3 may coregulate flower development with three flower-specific MADS-domain TFs and four basic helix-loop-helix TFs that are involved in photomorphogenesis. We further demonstrate that CLV3, SEPALLATA1 (SEP1), and SEP2 are FHY3 target genes. In shoot apical meristem, FHY3 directly represses CLV3, which consequently regulates WUS to maintain the stem cell pool. Intriguingly, CLV3 expression did not change significantly in fhy3 and phytochrome B mutants before and after light treatment, indicating that FHY3 and phytochrome B are involved in light-regulated meristem activity. In FM, FHY3 directly represses CLV3, but activates SEP2, to ultimately promote FM determinacy. Taken together, our results reveal insights into the mechanisms of meristem maintenance and determinacy, and illustrate how the roles of a single TF may vary in different organs and developmental stages.
Keywords: CLV3; FHY3; SEP2; meristem determinacy; meristem maintenance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Sablowski R. Flowering and determinacy in Arabidopsis. J Exp Bot. 2007;58(5):899–907. - PubMed
-
- Laux T, Mayer KFX, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122(1):87–96. - PubMed
-
- Mayer KFX, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. - PubMed
-
- Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

