Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-Hydroxyphenazine
- PMID: 27470070
- PMCID: PMC4965901
- DOI: 10.1186/s12934-016-0529-0
Genetic engineering of Pseudomonas chlororaphis GP72 for the enhanced production of 2-Hydroxyphenazine
Abstract
Background: The biocontrol strain Pseudomonas chlororaphis GP72 isolated from the green pepper rhizosphere synthesizes three antifungal phenazine compounds, 2-Hydroxyphenazine (2-OH-PHZ), 2-hydroxy-phenazine-1-carboxylic acid (2-OH-PCA) and phenazine-1-carboxylic acid (PCA). PCA has been a commercialized antifungal pesticide registered as "Shenqinmycin" in China since 2011. It is found that 2-OH-PHZ shows stronger fungistatic and bacteriostatic activity to some pathogens than PCA. 2-OH-PHZ could be developed as a potential antifungal pesticide. But the yield of 2-OH-PHZ generally is quite low, such as P. chlororaphis GP72, the production of 2-OH-PHZ by the wide-type strain is only 4.5 mg/L, it is necessary to enhance the yield of 2-OH-PHZ for its application in agriculture.
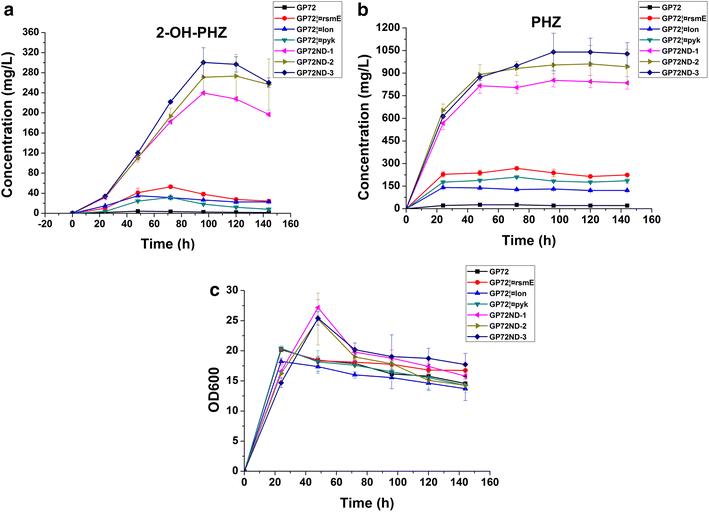
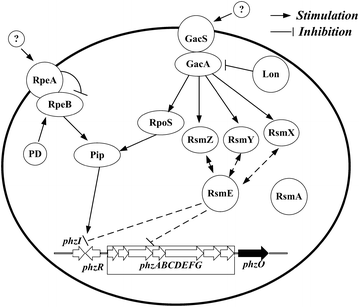
Results: Different strategies were used to improve the yield of 2-OH-PHZ: knocking out the negative regulatory genes, enhancing the shikimate pathway, deleting the competing pathways of 2-OH-PHZ synthesis based on chorismate, and improving the activity of PhzO which catalyzes the conversion of PCA to 2-OH-PHZ, although the last two strategies did not give us satisfactory results. In this study, four negative regulatory genes (pykF, rpeA, rsmE and lon) were firstly knocked out of the strain GP72 genome stepwise. The yield of 2-OH-PHZ improved more than 60 folds and increased from 4.5 to about 300 mg/L. Then six key genes (ppsA, tktA, phzC, aroB, aroD and aroE) selected from the gluconeogenesis, pentose phosphate and shikimate pathways which used to enhance the shikimate pathway were overexpressed to improve the production of 2-OH-PHZ. At last a genetically engineered strain that increased the 2-OH-PHZ production by 99-fold to 450.4 mg/L was obtained.
Conclusions: The 2-OH-PHZ production of P. chlororaphis GP72 was greatly improved through disruption of four negative regulatory genes and overexpression of six key genes, and it is shown that P. chlororaphis GP72 could be modified as a potential cell factory to produce 2-OH-PHZ and other phenazine biopesticides by genetic and metabolic engineering.
Keywords: 2-Hydroxyphenazine; Non-scar deletion; Overexpression; Phenazine-1-carboxylic acid; Pseudomonas chlororaphis GP72.
Figures



Similar articles
-
Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72.Appl Microbiol Biotechnol. 2011 Jan;89(1):169-77. doi: 10.1007/s00253-010-2863-1. Epub 2010 Sep 21. Appl Microbiol Biotechnol. 2011. PMID: 20857290
-
Developing genome-reduced Pseudomonas chlororaphis strains for the production of secondary metabolites.BMC Genomics. 2017 Sep 11;18(1):715. doi: 10.1186/s12864-017-4127-2. BMC Genomics. 2017. PMID: 28893188 Free PMC article.
-
Comparative metabolomics and transcriptomics analyses provide insights into the high-yield mechanism of phenazines biosynthesis in Pseudomonas chlororaphis GP72.J Appl Microbiol. 2022 Nov;133(5):2790-2801. doi: 10.1111/jam.15727. Epub 2022 Aug 5. J Appl Microbiol. 2022. PMID: 35870153
-
Insights into plant-beneficial traits of probiotic Pseudomonas chlororaphis isolates.J Med Microbiol. 2020 Mar;69(3):361-371. doi: 10.1099/jmm.0.001157. J Med Microbiol. 2020. PMID: 32043956 Review.
-
Pseudomonas chlororaphis metabolites as biocontrol promoters of plant health and improved crop yield.World J Microbiol Biotechnol. 2021 May 12;37(6):99. doi: 10.1007/s11274-021-03063-w. World J Microbiol Biotechnol. 2021. PMID: 33978868 Review.
Cited by
-
Understanding the Contribution of Lactate Metabolism in Cancer Progress: A Perspective from Isomers.Cancers (Basel). 2022 Dec 23;15(1):87. doi: 10.3390/cancers15010087. Cancers (Basel). 2022. PMID: 36612084 Free PMC article. Review.
-
Screening of natural phenazine producers for electroactivity in bioelectrochemical systems.Microb Biotechnol. 2023 Mar;16(3):579-594. doi: 10.1111/1751-7915.14199. Epub 2022 Dec 26. Microb Biotechnol. 2023. PMID: 36571174 Free PMC article.
-
Biosynthesis and metabolic engineering of 1-hydroxyphenazine in Pseudomonas chlororaphis H18.Microb Cell Fact. 2021 Dec 30;20(1):235. doi: 10.1186/s12934-021-01731-y. Microb Cell Fact. 2021. PMID: 34965873 Free PMC article.
-
Engineering and systems-level analysis of Pseudomonas chlororaphis for production of phenazine-1-carboxamide using glycerol as the cost-effective carbon source.Biotechnol Biofuels. 2018 May 4;11:130. doi: 10.1186/s13068-018-1123-y. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29755589 Free PMC article.
-
Root-Associated Bacteria Are Biocontrol Agents for Multiple Plant Pests.Microorganisms. 2022 May 19;10(5):1053. doi: 10.3390/microorganisms10051053. Microorganisms. 2022. PMID: 35630495 Free PMC article. Review.
References
-
- Du X, Li Y, Zhou W, Zhou Q, Liu H, Xu Y. Phenazine-1-carboxylic acid production in a chromosomally non-scar triple-deleted mutant Pseudomonas aeruginosa using statistical experimental designs to optimize yield. Appl Microbiol Biotechnol. 2013;97:7767–7778. doi: 10.1007/s00253-013-4921-y. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

