Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin
- PMID: 27470111
- PMCID: PMC5052058
- DOI: 10.1101/gr.209106.116
Global genome nucleotide excision repair is organized into domains that promote efficient DNA repair in chromatin
Abstract
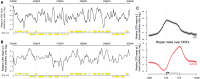
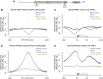
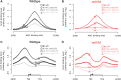
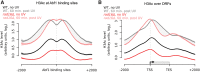
The rates at which lesions are removed by DNA repair can vary widely throughout the genome, with important implications for genomic stability. To study this, we measured the distribution of nucleotide excision repair (NER) rates for UV-induced lesions throughout the budding yeast genome. By plotting these repair rates in relation to genes and their associated flanking sequences, we reveal that, in normal cells, genomic repair rates display a distinctive pattern, suggesting that DNA repair is highly organized within the genome. Furthermore, by comparing genome-wide DNA repair rates in wild-type cells and cells defective in the global genome-NER (GG-NER) subpathway, we establish how this alters the distribution of NER rates throughout the genome. We also examined the genomic locations of GG-NER factor binding to chromatin before and after UV irradiation, revealing that GG-NER is organized and initiated from specific genomic locations. At these sites, chromatin occupancy of the histone acetyl-transferase Gcn5 is controlled by the GG-NER complex, which regulates histone H3 acetylation and chromatin structure, thereby promoting efficient DNA repair of UV-induced lesions. Chromatin remodeling during the GG-NER process is therefore organized into these genomic domains. Importantly, loss of Gcn5 significantly alters the genomic distribution of NER rates; this has implications for the effects of chromatin modifiers on the distribution of mutations that arise throughout the genome.
© 2016 Yu et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae.PLoS Genet. 2011 Jun;7(6):e1002124. doi: 10.1371/journal.pgen.1002124. Epub 2011 Jun 16. PLoS Genet. 2011. PMID: 21698136 Free PMC article.
-
Histone modification and chromatin remodeling during NER.DNA Repair (Amst). 2015 Dec;36:105-113. doi: 10.1016/j.dnarep.2015.09.013. Epub 2015 Sep 16. DNA Repair (Amst). 2015. PMID: 26422133 Review.
-
Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes.Nucleic Acids Res. 2013 Oct;41(19):9006-19. doi: 10.1093/nar/gkt688. Epub 2013 Aug 7. Nucleic Acids Res. 2013. PMID: 23925126 Free PMC article.
-
Nucleotide excision repair in cellular chromatin: studies with yeast from nucleotide to gene to genome.Int J Mol Sci. 2012;13(9):11141-11164. doi: 10.3390/ijms130911141. Epub 2012 Sep 7. Int J Mol Sci. 2012. PMID: 23109843 Free PMC article. Review.
-
NuA4 acetyltransferase is required for efficient nucleotide excision repair in yeast.DNA Repair (Amst). 2019 Jan;73:91-98. doi: 10.1016/j.dnarep.2018.11.006. Epub 2018 Nov 14. DNA Repair (Amst). 2019. PMID: 30473425 Free PMC article.
Cited by
-
Population sequencing data reveal a compendium of mutational processes in the human germ line.Science. 2021 Aug 27;373(6558):1030-1035. doi: 10.1126/science.aba7408. Epub 2021 Aug 12. Science. 2021. PMID: 34385354 Free PMC article.
-
Nucleosome remodeling at origins of global genome-nucleotide excision repair occurs at the boundaries of higher-order chromatin structure.Genome Res. 2019 Jan;29(1):74-84. doi: 10.1101/gr.237198.118. Epub 2018 Dec 14. Genome Res. 2019. PMID: 30552104 Free PMC article.
-
Histone variant H2A.Z is needed for efficient transcription-coupled NER and genome integrity in UV challenged yeast cells.PLoS Genet. 2024 Sep 10;20(9):e1011300. doi: 10.1371/journal.pgen.1011300. eCollection 2024 Sep. PLoS Genet. 2024. PMID: 39255275 Free PMC article.
-
Mechanisms of radiation-induced tissue damage and response.MedComm (2020). 2024 Sep 20;5(10):e725. doi: 10.1002/mco2.725. eCollection 2024 Oct. MedComm (2020). 2024. PMID: 39309694 Free PMC article. Review.
-
Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):11507-11512. doi: 10.1073/pnas.1614430113. Epub 2016 Sep 29. Proc Natl Acad Sci U S A. 2016. PMID: 27688757 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
