Molecular Basis of Chemokine CXCL5-Glycosaminoglycan Interactions
- PMID: 27471273
- PMCID: PMC5034048
- DOI: 10.1074/jbc.M116.745265
Molecular Basis of Chemokine CXCL5-Glycosaminoglycan Interactions
Abstract
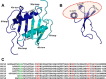
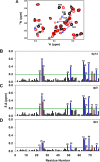
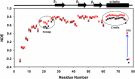
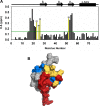
Chemokines, a large family of highly versatile small soluble proteins, play crucial roles in defining innate and adaptive immune responses by regulating the trafficking of leukocytes, and also play a key role in various aspects of human physiology. Chemokines share the characteristic feature of reversibly existing as monomers and dimers, and their functional response is intimately coupled to interaction with glycosaminoglycans (GAGs). Currently, nothing is known regarding the structural basis or molecular mechanisms underlying CXCL5-GAG interactions. To address this missing knowledge, we characterized the interaction of a panel of heparin oligosaccharides to CXCL5 using solution NMR, isothermal titration calorimetry, and molecular dynamics simulations. NMR studies indicated that the dimer is the high-affinity GAG binding ligand and that lysine residues from the N-loop, 40s turn, β3 strand, and C-terminal helix mediate binding. Isothermal titration calorimetry indicated a stoichiometry of two oligosaccharides per CXCL5 dimer. NMR-based structural models reveal that these residues form a contiguous surface within a monomer and, interestingly, that the GAG-binding domain overlaps with the receptor-binding domain, indicating that a GAG-bound chemokine cannot activate the receptor. Molecular dynamics simulations indicate that the roles of the individual lysines are not equivalent and that helical lysines play a more prominent role in determining binding geometry and affinity. Further, binding interactions and GAG geometry in CXCL5 are novel and distinctly different compared with the related chemokines CXCL1 and CXCL8. We conclude that a finely tuned balance between the GAG-bound dimer and free soluble monomer regulates CXCL5-mediated receptor signaling and function.
Keywords: NMR; glycobiology; heparin; isothermal titration calorimetry (ITC); molecular dynamics.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Bonecchi R., Galliera E., Borroni E. M., Corsi M. M., Locati M., and Mantovani A. (2009) Chemokines and chemokine receptors: an overview. Front. Biosci. 14, 540–551 - PubMed
-
- Griffith J. W., Sokol C. L., and Luster A. D. (2014) Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 - PubMed
-
- Fernandez E. J., and Lolis E. (2002) Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 42, 469–499 - PubMed
-
- Zlotnik A., and Yoshie O. (2000) Chemokines: a new classification system and their role in immunity. Immunity 12, 121–127 - PubMed
-
- Stillie R., Farooq S. M., Gordon J. R., and Stadnyk A. W. (2009) The functional significance behind expressing two IL-8 receptor types on PMN. J. Leukocyte Biol. 86, 529–543 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

