The Use of Multiscale Molecular Simulations in Understanding a Relationship between the Structure and Function of Biological Systems of the Brain: The Application to Monoamine Oxidase Enzymes
- PMID: 27471444
- PMCID: PMC4945635
- DOI: 10.3389/fnins.2016.00327
The Use of Multiscale Molecular Simulations in Understanding a Relationship between the Structure and Function of Biological Systems of the Brain: The Application to Monoamine Oxidase Enzymes
Abstract
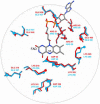
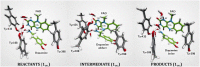
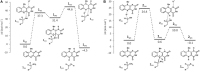
HIGHLIGHTS Computational techniques provide accurate descriptions of the structure and dynamics of biological systems, contributing to their understanding at an atomic level.Classical MD simulations are a precious computational tool for the processes where no chemical reactions take place.QM calculations provide valuable information about the enzyme activity, being able to distinguish among several mechanistic pathways, provided a carefully selected cluster model of the enzyme is considered.Multiscale QM/MM simulation is the method of choice for the computational treatment of enzyme reactions offering quantitative agreement with experimentally determined reaction parameters.Molecular simulation provide insight into the mechanism of both the catalytic activity and inhibition of monoamine oxidases, thus aiding in the rational design of their inhibitors that are all employed and antidepressants and antiparkinsonian drugs. Aging society and therewith associated neurodegenerative and neuropsychiatric diseases, including depression, Alzheimer's disease, obsessive disorders, and Parkinson's disease, urgently require novel drug candidates. Targets include monoamine oxidases A and B (MAOs), acetylcholinesterase (AChE), butyrylcholinesterase (BChE), and various receptors and transporters. For rational drug design it is particularly important to combine experimental synthetic, kinetic, toxicological, and pharmacological information with structural and computational work. This paper describes the application of various modern computational biochemistry methods in order to improve the understanding of a relationship between the structure and function of large biological systems including ion channels, transporters, receptors, and metabolic enzymes. The methods covered stem from classical molecular dynamics simulations to understand the physical basis and the time evolution of the structures, to combined QM, and QM/MM approaches to probe the chemical mechanisms of enzymatic activities and their inhibition. As an illustrative example, the later will focus on the monoamine oxidase family of enzymes, which catalyze the degradation of amine neurotransmitters in various parts of the brain, the imbalance of which is associated with the development and progression of a range of neurodegenerative disorders. Inhibitors that act mainly on MAO A are used in the treatment of depression, due to their ability to raise serotonin concentrations, while MAO B inhibitors decrease dopamine degradation and improve motor control in patients with Parkinson disease. Our results give strong support that both MAO isoforms, A and B, operate through the hydride transfer mechanism. Relevance of MAO catalyzed reactions and MAO inhibition in the context of neurodegeneration will be discussed.
Keywords: central nervous system; computational enzymology; drug design; hydride transfer reaction; molecular dynamics simulation; multiscale simulations; neural signal transduction; neurotransmitter metabolism.
Figures


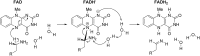

Similar articles
-
Computational Insight into the Mechanism of the Irreversible Inhibition of Monoamine Oxidase Enzymes by the Antiparkinsonian Propargylamine Inhibitors Rasagiline and Selegiline.ACS Chem Neurosci. 2019 Aug 21;10(8):3532-3542. doi: 10.1021/acschemneuro.9b00147. Epub 2019 Jul 2. ACS Chem Neurosci. 2019. PMID: 31264403
-
ONIOM calculations on serotonin degradation by monoamine oxidase B: insight into the oxidation mechanism and covalent reversible inhibition.Org Biomol Chem. 2016 Oct 21;14(39):9239-9252. doi: 10.1039/c6ob01175f. Epub 2016 Sep 7. Org Biomol Chem. 2016. PMID: 27605388
-
Hydride Abstraction as the Rate-Limiting Step of the Irreversible Inhibition of Monoamine Oxidase B by Rasagiline and Selegiline: A Computational Empirical Valence Bond Study.Int J Mol Sci. 2020 Aug 26;21(17):6151. doi: 10.3390/ijms21176151. Int J Mol Sci. 2020. PMID: 32858935 Free PMC article.
-
Monoamine Oxidase Inhibitors: From Classic to New Clinical Approaches.Handb Exp Pharmacol. 2021;264:229-259. doi: 10.1007/164_2020_384. Handb Exp Pharmacol. 2021. PMID: 32852645 Review.
-
Recent Advancements and SAR Studies of Synthetic Coumarins as MAO-B Inhibitors: An Updated Review.Mini Rev Med Chem. 2024;24(20):1834-1846. doi: 10.2174/0113895575290599240503080025. Mini Rev Med Chem. 2024. PMID: 38778598 Review.
Cited by
-
Drug Development for Alzheimer's and Parkinson's Disease: Where Do We Go Now?Pharmaceutics. 2024 May 24;16(6):708. doi: 10.3390/pharmaceutics16060708. Pharmaceutics. 2024. PMID: 38931832 Free PMC article. Review.
-
Computational Mechanistic Study of l-Aspartate Oxidase by ONIOM Method.ACS Omega. 2023 May 25;8(22):19963-19968. doi: 10.1021/acsomega.3c01949. eCollection 2023 Jun 6. ACS Omega. 2023. PMID: 37305300 Free PMC article.
-
Mechanistic Understanding from Molecular Dynamics in Pharmaceutical Research 2: Lipid Membrane in Drug Design.Pharmaceuticals (Basel). 2021 Oct 19;14(10):1062. doi: 10.3390/ph14101062. Pharmaceuticals (Basel). 2021. PMID: 34681286 Free PMC article. Review.
-
Empirical Valence Bond Simulations Suggest a Direct Hydride Transfer Mechanism for Human Diamine Oxidase.ACS Omega. 2018 Apr 30;3(4):3665-3674. doi: 10.1021/acsomega.8b00346. Epub 2018 Apr 2. ACS Omega. 2018. PMID: 30023875 Free PMC article.
-
Amphetamine Derivatives as Monoamine Oxidase Inhibitors.Front Pharmacol. 2020 Jan 23;10:1590. doi: 10.3389/fphar.2019.01590. eCollection 2019. Front Pharmacol. 2020. PMID: 32038257 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

