Corynoline Isolated from Corydalis bungeana Turcz. Exhibits Anti-Inflammatory Effects via Modulation of Nfr2 and MAPKs
- PMID: 27472313
- PMCID: PMC6273489
- DOI: 10.3390/molecules21080975
Corynoline Isolated from Corydalis bungeana Turcz. Exhibits Anti-Inflammatory Effects via Modulation of Nfr2 and MAPKs
Abstract
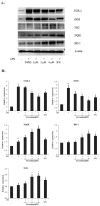
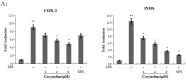
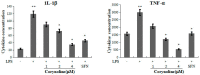
Corydalis bungeana Turcz. is an anti-inflammatory medicinal herb used widely in traditional Chinese medicine for upper respiratory tract infections. It is demonstrated that corynoline is its active anti-inflammatory component. The nuclear factor-erythroid-2-related factor 2 (Nrf2)/antioxidant response element (ARE) pathway and the mitogen-activated protein kinase (MAPK) pathway play important roles in the regulation of inflammation. In this study, we investigated the potential anti-inflammatory mechanism of corynoline through modulation of Nfr2 and MAPKs. Lipopolysaccharide (LPS)-activated RAW264.7 cells were used to explore modulatory role of NO production and the activation of signaling proteins and transcription factors using nitrite assay, Western bloting and qPCR. Treatment with corynoline reduced production of nitric oxide (NO) and the protein and mRNA levels of inducible nitric oxide (iNOS) and cyclooxygenase-2 (COX-2) Treatment also significantly increased the expression of Nrf2, quinone oxidoreductase 1 (NQO1) and hemeoxygenase-1 (HO-1) at the mRNA and protein levels, which demonstrated that corynoline may protect cells from inflammation through the Nrf2/ARE pathway In addition, corynoline suppressed the expression of inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), at the mRNA and protein levels. Furthermore, molecular data revealed that corynoline inhibited lipopolysaccharide-stimulated phosphorylation of c-jun NH2-terminal kinase (JNK) and p38. Taken together, these results suggest that corynoline reduces the levels of pro-inflammatory mediators, such as iNOS, COX-2, TNF-α and IL-1β, by suppressing extracellular signal-regulated kinase 1/2 (ERK) and p38 phosphorylation in RAW264.7 cells, which is regulated by the Nrf2/ARE pathway. These findings reveal part of the molecular basis for the anti-inflammatory properties of corynoline.
Keywords: MAPKs; Nrf2; anti-inflammation; corynoline.
Conflict of interest statement
The authors do not have conflict of interest to declare.
Figures
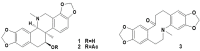
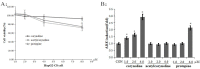




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

