The Complete Genome Sequence of Plodia Interpunctella Granulovirus: Evidence for Horizontal Gene Transfer and Discovery of an Unusual Inhibitor-of-Apoptosis Gene
- PMID: 27472489
- PMCID: PMC4966970
- DOI: 10.1371/journal.pone.0160389
The Complete Genome Sequence of Plodia Interpunctella Granulovirus: Evidence for Horizontal Gene Transfer and Discovery of an Unusual Inhibitor-of-Apoptosis Gene
Abstract
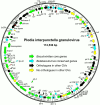
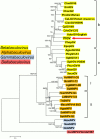
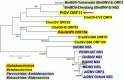
The Indianmeal moth, Plodia interpunctella (Lepidoptera: Pyralidae), is a common pest of stored goods with a worldwide distribution. The complete genome sequence for a larval pathogen of this moth, the baculovirus Plodia interpunctella granulovirus (PiGV), was determined by next-generation sequencing. The PiGV genome was found to be 112, 536 bp in length with a 44.2% G+C nucleotide distribution. A total of 123 open reading frames (ORFs) and seven homologous regions (hrs) were identified and annotated. Phylogenetic inference using concatenated alignments of 36 baculovirus core genes placed PiGV in the "b" clade of viruses from genus Betabaculovirus with a branch length suggesting that PiGV represents a distinct betabaculovirus species. In addition to the baculovirus core genes and orthologues of other genes found in other betabaculovirus genomes, the PiGV genome sequence contained orthologues of the bidensovirus NS3 gene, as well as ORFs that occur in alphabaculoviruses but not betabaculoviruses. While PiGV contained an orthologue of inhibitor of apoptosis-5 (iap-5), an orthologue of inhibitor of apoptosis-3 (iap-3) was not present. Instead, the PiGV sequence contained an ORF (PiGV ORF81) encoding an IAP homologue with sequence similarity to insect cellular IAPs, but not to viral IAPs. Phylogenetic analysis of baculovirus and insect IAP amino acid sequences suggested that the baculovirus IAP-3 genes and the PiGV ORF81 IAP homologue represent different lineages arising from more than one acquisition event. The presence of genes from other sources in the PiGV genome highlights the extent to which baculovirus gene content is shaped by horizontal gene transfer.
Conflict of interest statement
Figures





Similar articles
-
The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta.PLoS One. 2017 Jan 19;12(1):e0170510. doi: 10.1371/journal.pone.0170510. eCollection 2017. PLoS One. 2017. PMID: 28103323 Free PMC article.
-
Genome of Cnaphalocrocis medinalis Granulovirus, the First Crambidae-Infecting Betabaculovirus Isolated from Rice Leaffolder to Sequenced.PLoS One. 2016 Feb 5;11(2):e0147882. doi: 10.1371/journal.pone.0147882. eCollection 2016. PLoS One. 2016. PMID: 26848752 Free PMC article.
-
A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes.Viruses. 2018 Mar 16;10(3):134. doi: 10.3390/v10030134. Viruses. 2018. PMID: 29547534 Free PMC article.
-
Inhibitor of apoptosis proteins in eukaryotic evolution and development: a model of thematic conservation.Dev Cell. 2008 Oct;15(4):497-508. doi: 10.1016/j.devcel.2008.09.012. Dev Cell. 2008. PMID: 18854135 Free PMC article. Review.
-
The conservation of IAP-like proteins in fungi, and their potential role in fungal programmed cell death.Fungal Genet Biol. 2022 Sep;162:103730. doi: 10.1016/j.fgb.2022.103730. Epub 2022 Aug 23. Fungal Genet Biol. 2022. PMID: 35998750 Review.
Cited by
-
Regulatory Mechanisms, Protein Expression and Biological Activity of Photolyase Gene from Spodoptera littoralis Granulovirus Genome.Mol Biotechnol. 2023 Mar;65(3):433-440. doi: 10.1007/s12033-022-00537-6. Epub 2022 Aug 18. Mol Biotechnol. 2023. PMID: 35980593 Free PMC article.
-
Genome Analysis of a Novel Clade b Betabaculovirus Isolated from the Legume Pest Matsumuraeses phaseoli (Lepidoptera: Tortricidae).Viruses. 2020 Sep 23;12(10):1068. doi: 10.3390/v12101068. Viruses. 2020. PMID: 32977681 Free PMC article.
-
The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta.PLoS One. 2017 Jan 19;12(1):e0170510. doi: 10.1371/journal.pone.0170510. eCollection 2017. PLoS One. 2017. PMID: 28103323 Free PMC article.
-
The Genome Sequences of Baculoviruses from the Tufted Apple Bud Moth, Platynota idaeusalis, Reveal Recombination Between an Alphabaculovirus and a Betabaculovirus from the Same Host.Viruses. 2025 Jan 30;17(2):202. doi: 10.3390/v17020202. Viruses. 2025. PMID: 40006957 Free PMC article.
-
Characterization of the RNA Transcription Profile of Bombyx mori Bidensovirus.Viruses. 2019 Apr 3;11(4):325. doi: 10.3390/v11040325. Viruses. 2019. PMID: 30987230 Free PMC article.
References
-
- Harrison RL, Hoover K. Baculoviruses and Other Occluded Insect Viruses In: Vega FE, Kaya HK, editors. Insect Pathology, Second Edition Boston: Academic Press; 2012. p. 73–131.
-
- Herniou EA, Arif BM, Becnel JJ, Blissard GW, Bonning B, Harrison RL, et al. Baculoviridae In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Oxford: Elsevier; 2012. p. 163–174.
-
- Winstanley D, Crook NE. Replication of Cydia pomonella granulosis virus in cell cultures. J Gen Virol. 1993; 74:1599–1609. - PubMed
-
- Arnott HJ, Smith KM. An ultrastructural study of the development of a granulosis virus in the cells of the moth Plodia interpunctella. Journal of Ultrastructure Research. 1968;21:251–268. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

