Evidence of selection for an accessible nucleosomal array in human
- PMID: 27472913
- PMCID: PMC4966569
- DOI: 10.1186/s12864-016-2880-2
Evidence of selection for an accessible nucleosomal array in human
Abstract
Background: Recently, a physical model of nucleosome formation based on sequence-dependent bending properties of the DNA double-helix has been used to reveal some enrichment of nucleosome-inhibiting energy barriers (NIEBs) nearby ubiquitous human "master" replication origins. Here we use this model to predict the existence of about 1.6 millions NIEBs over the 22 human autosomes.
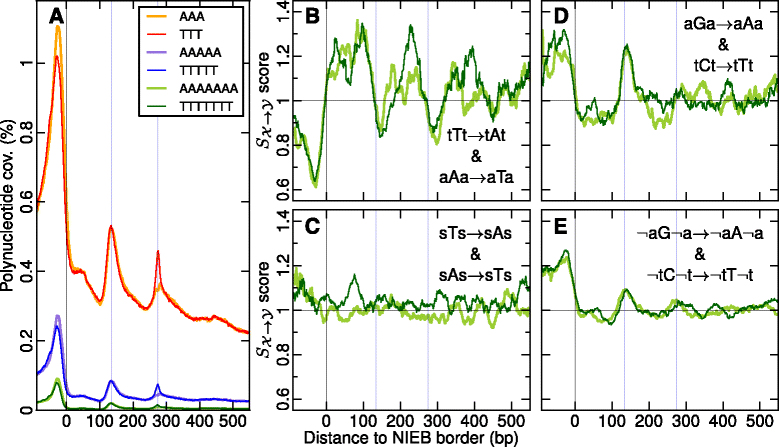
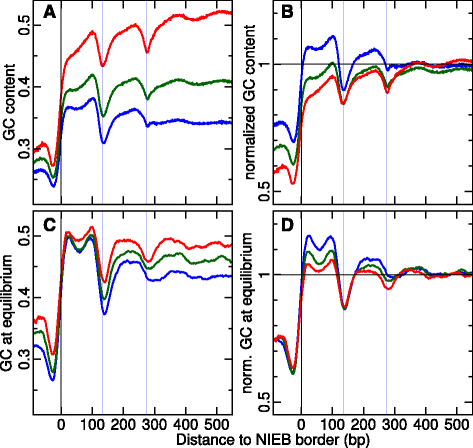
Results: We show that these high energy barriers of mean size 153 bp correspond to nucleosome-depleted regions (NDRs) in vitro, as expected, but also in vivo. On either side of these NIEBs, we observe, in vivo and in vitro, a similar compacted nucleosome ordering, suggesting an absence of chromatin remodeling. This nucleosomal ordering strongly correlates with oscillations of the GC content as well as with the interspecies and intraspecies mutation profiles along these regions. Comparison of these divergence rates reveals the existence of both positive and negative selections linked to nucleosome positioning around these intrinsic NDRs. Overall, these NIEBs and neighboring nucleosomes cover 37.5 % of the human genome where nucleosome occupancy is stably encoded in the DNA sequence. These 1 kb-sized regions of intrinsic nucleosome positioning are equally found in GC-rich and GC-poor isochores, in early and late replicating regions, in intergenic and genic regions but not at gene promoters.
Conclusion: The source of selection pressure on the NIEBs has yet to be resolved in future work. One possible scenario is that these widely distributed chromatin patterns have been selected in human to impair the condensation of the nucleosomal array into the 30 nm chromatin fiber, so as to facilitate the epigenetic regulation of nuclear functions in a cell-type-specific manner.
Keywords: GC content; Human genome; Mutation rates; Nucleosome depleted regions; Nucleosome ordering; Sequence evolution.
Figures

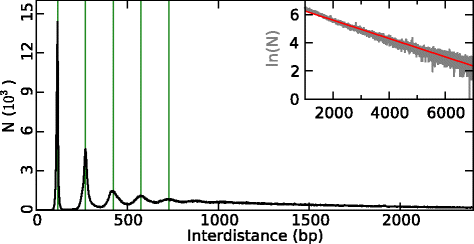
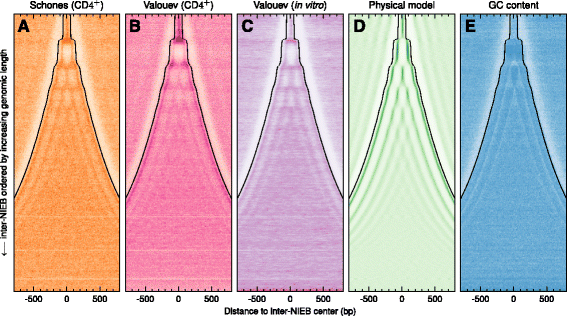
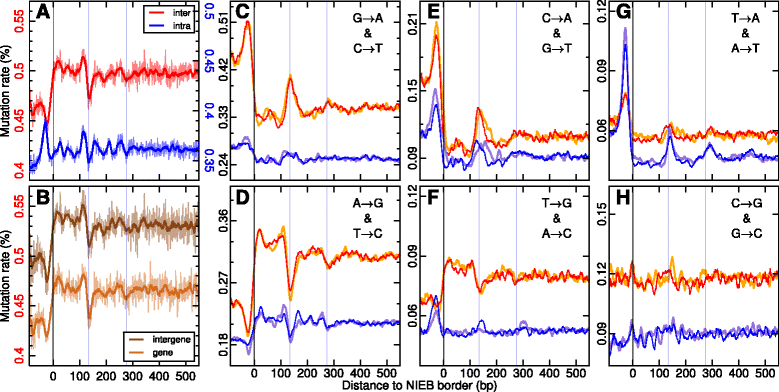
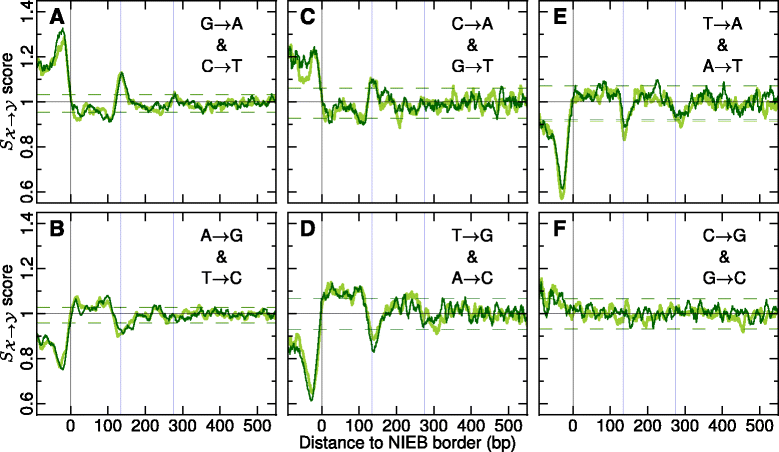
References
-
- Arneodo A, Vaillant C, Audit B, Argoul F, d’Aubenton-Carafa Y, Thermes C. Multi-scale coding of genomic information: From DNA sequence to genome structure and function. Phys Rep. 2011;498:45–188. doi: 10.1016/j.physrep.2010.10.001. - DOI
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. New-York: Garland Publishing; 2002.
-
- Graur D, Li WH. Fundamentals of Molecular Evolution. Sunderland: Sinauer Associates; 1999.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

