Identification of combinatorial host-specific signatures with a potential to affect host adaptation in influenza A H1N1 and H3N2 subtypes
- PMID: 27473048
- PMCID: PMC4966792
- DOI: 10.1186/s12864-016-2919-4
Identification of combinatorial host-specific signatures with a potential to affect host adaptation in influenza A H1N1 and H3N2 subtypes
Abstract
Background: The underlying strategies used by influenza A viruses (IAVs) to adapt to new hosts while crossing the species barrier are complex and yet to be understood completely. Several studies have been published identifying singular genomic signatures that indicate such a host switch. The complexity of the problem suggested that in addition to the singular signatures, there might be a combinatorial use of such genomic features, in nature, defining adaptation to hosts.
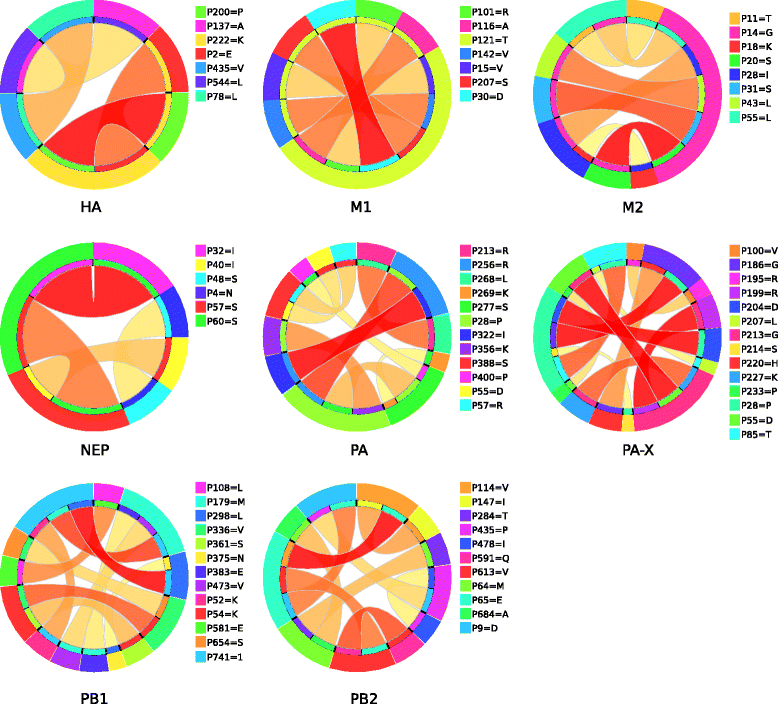
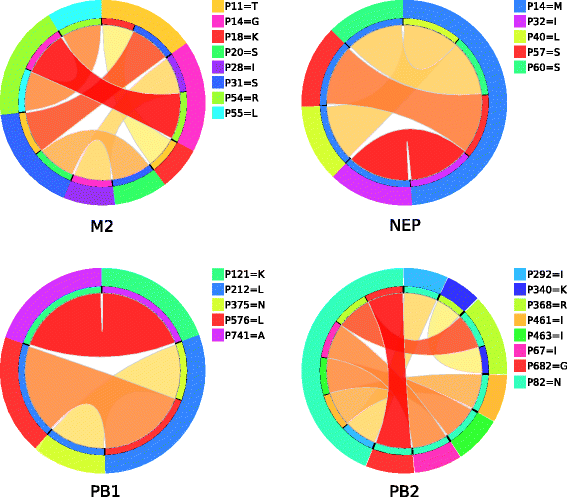
Results: We used computational rule-based modeling to identify combinatorial sets of interacting amino acid (aa) residues in 12 proteins of IAVs of H1N1 and H3N2 subtypes. We built highly accurate rule-based models for each protein that could differentiate between viral aa sequences coming from avian and human hosts. We found 68 host-specific combinations of aa residues, potentially associated to host adaptation on HA, M1, M2, NP, NS1, NEP, PA, PA-X, PB1 and PB2 proteins of the H1N1 subtype and 24 on M1, M2, NEP, PB1 and PB2 proteins of the H3N2 subtypes. In addition to these combinations, we found 132 novel singular aa signatures distributed among all proteins, including the newly discovered PA-X protein, of both subtypes. We showed that HA, NA, NP, NS1, NEP, PA-X and PA proteins of the H1N1 subtype carry H1N1-specific and HA, NA, PA-X, PA, PB1-F2 and PB1 of the H3N2 subtype carry H3N2-specific signatures. M1, M2, PB1-F2, PB1 and PB2 of H1N1 subtype, in addition to H1N1 signatures, also carry H3N2 signatures. Similarly M1, M2, NP, NS1, NEP and PB2 of H3N2 subtype were shown to carry both H3N2 and H1N1 host-specific signatures (HSSs).
Conclusions: To sum it up, we computationally constructed simple IF-THEN rule-based models that could distinguish between aa sequences of avian and human IAVs. From the rules we identified HSSs having a potential to affect the adaptation to specific hosts. The identification of combinatorial HSSs suggests that the process of adaptation of IAVs to a new host is more complex than previously suggested. The present study provides a basis for further detailed studies with the aim to elucidate the molecular mechanisms providing the foundation for the adaptation process.
Keywords: Combinatorial signatures; Host adaptation; Host-specific signatures; Influenza A virus; MCFS; Rosetta; Rough sets.
Figures

Similar articles
-
Surveillance in Eastern India (2007-2009) revealed reassortment event involving NS and PB1-F2 gene segments among co-circulating influenza A subtypes.Virol J. 2012 Jan 5;9:3. doi: 10.1186/1743-422X-9-3. Virol J. 2012. PMID: 22217077 Free PMC article.
-
Zoonotic Risk, Pathogenesis, and Transmission of Avian-Origin H3N2 Canine Influenza Virus.J Virol. 2017 Oct 13;91(21):e00637-17. doi: 10.1128/JVI.00637-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28814512 Free PMC article.
-
Seasonal H3N2 and 2009 Pandemic H1N1 Influenza A Viruses Reassort Efficiently but Produce Attenuated Progeny.J Virol. 2017 Aug 10;91(17):e00830-17. doi: 10.1128/JVI.00830-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637755 Free PMC article.
-
Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China.Biochem Biophys Res Commun. 2009 Aug 21;386(2):278-83. doi: 10.1016/j.bbrc.2009.05.056. Epub 2009 May 19. Biochem Biophys Res Commun. 2009. PMID: 19460353 Review.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
Cited by
-
Advantages of Broad-Spectrum Influenza mRNA Vaccines and Their Impact on Pulmonary Influenza.Vaccines (Basel). 2024 Dec 7;12(12):1382. doi: 10.3390/vaccines12121382. Vaccines (Basel). 2024. PMID: 39772044 Free PMC article. Review.
-
Misclassified: identification of zoonotic transition biomarker candidates for influenza A viruses using deep neural network.Front Genet. 2023 Jul 27;14:1145166. doi: 10.3389/fgene.2023.1145166. eCollection 2023. Front Genet. 2023. PMID: 37576548 Free PMC article.
-
Predicting Zoonotic Risk of Influenza A Viruses from Host Tropism Protein Signature Using Random Forest.Int J Mol Sci. 2017 May 25;18(6):1135. doi: 10.3390/ijms18061135. Int J Mol Sci. 2017. PMID: 28587080 Free PMC article.
-
Identifying host-specific amino acid signatures for influenza A viruses using an adjusted entropy measure.BMC Bioinformatics. 2022 Aug 12;23(1):333. doi: 10.1186/s12859-022-04885-7. BMC Bioinformatics. 2022. PMID: 35962315 Free PMC article.
-
Analysis of Expression Pattern of snoRNAs in Different Cancer Types with Machine Learning Algorithms.Int J Mol Sci. 2019 May 2;20(9):2185. doi: 10.3390/ijms20092185. Int J Mol Sci. 2019. PMID: 31052553 Free PMC article.
References
-
- Steel J, Lowen AC. Influenza A virus reassortment. Curr Top Microbiol Immunol. 2014;385:377–401. - PubMed
-
- cdc . Influenza (Seasonal) Fact Sheet. 2014.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

