Microbial diversity in individuals and their household contacts following typical antibiotic courses
- PMID: 27473422
- PMCID: PMC4967329
- DOI: 10.1186/s40168-016-0187-9
Microbial diversity in individuals and their household contacts following typical antibiotic courses
Abstract
Background: Antibiotics are a mainstay of treatment for bacterial infections worldwide, yet the effects of typical antibiotic prescriptions on human indigenous microbiota have not been thoroughly evaluated. We examined the effects of the two most commonly prescribed antibiotics (amoxicillin and azithromycin) in the USA to discern whether short-term antibiotic courses may have prolonged effects on human microbiota.
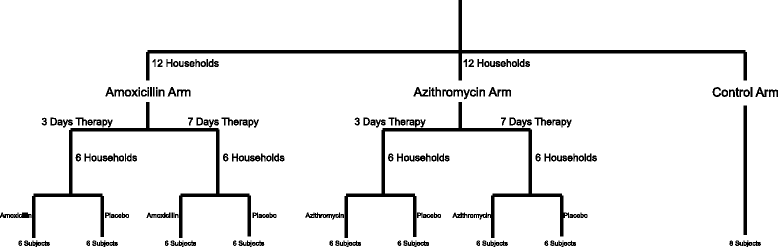
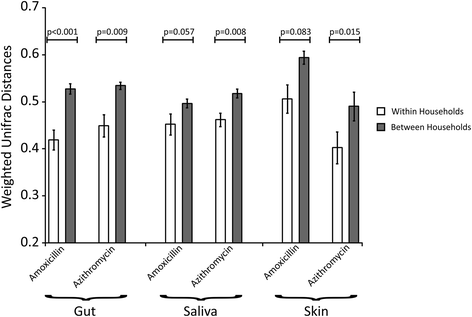
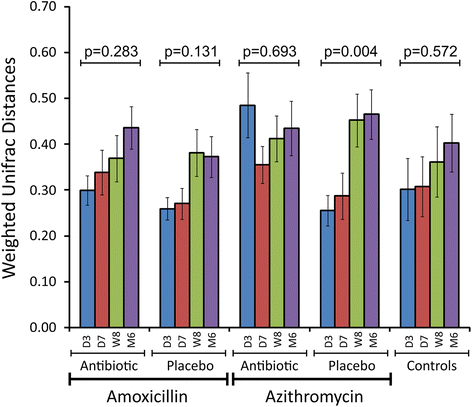
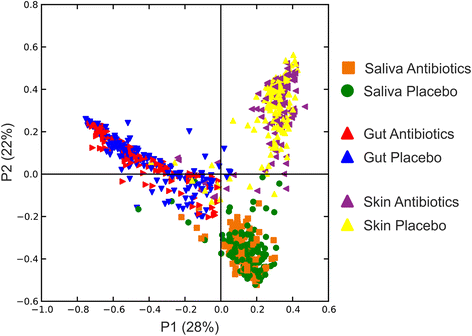
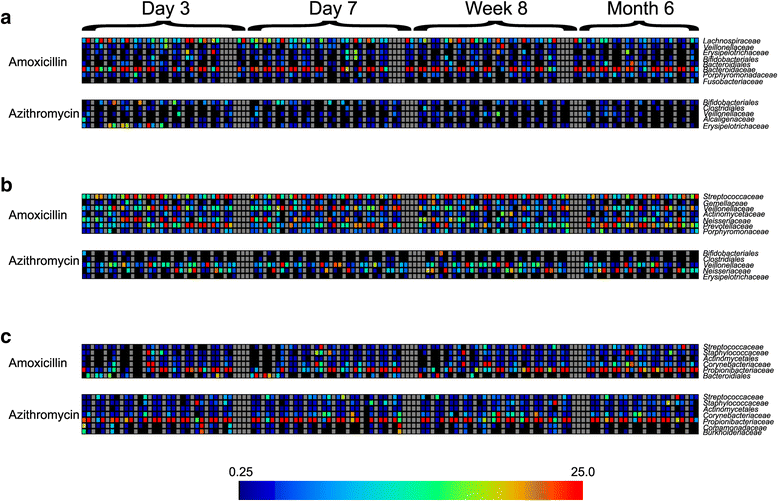
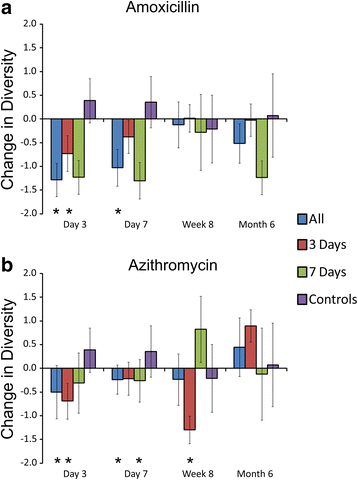
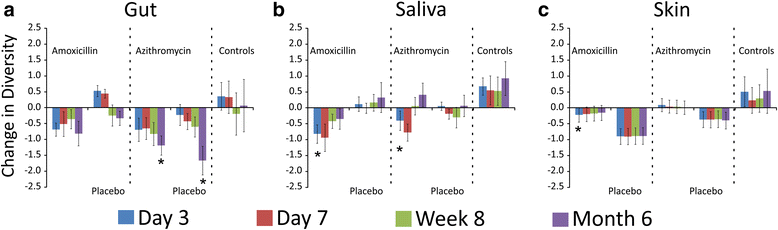
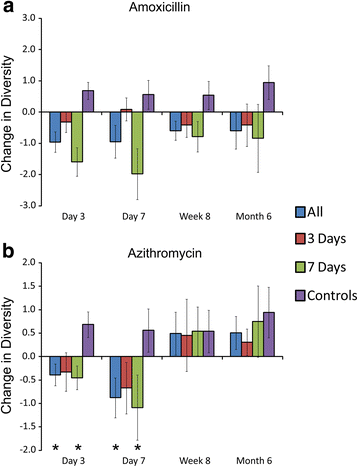
Results: We sampled the feces, saliva, and skin specimens from a cohort of unrelated, cohabitating individuals over 6 months. An individual in each household was given an antibiotic, and the other a placebo to discern antibiotic impacts on microbiota, as well as determine whether antibiotic use might reshape the microbiota of each household. We observed household-specific patterns of microbiota on each body surface, which persevered despite antibiotic perturbations. While the gut microbiota within an individual became more dissimilar over time, there was no evidence that the use of antibiotics accelerated this process when compared to household members. There was a significant change in microbiota diversity in the gut and mouth in response to antibiotics, but analogous patterns were not observed on the skin. Those who received 7 days of amoxicillin generally had greater reductions in diversity compared to those who received 3 days, in contrast to those who received azithromycin.
Conclusions: As few as 3 days of treatment with the most commonly prescribed antibiotics can result in sustained reductions in microbiota diversity, which could have implications for the maintenance of human health and resilience to disease.
Keywords: 16S rRNA; Antibiotic courses; Antibiotic perturbations; Antibiotics; Gut; Microbiome; Saliva; Skin.
Figures
References
-
- Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP, Jr, Rohwer F, Haynes M. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

