Clonal evolution of acute myeloid leukemia highlighted by latest genome sequencing studies
- PMID: 27474172
- PMCID: PMC5295455
- DOI: 10.18632/oncotarget.10850
Clonal evolution of acute myeloid leukemia highlighted by latest genome sequencing studies
Abstract
Decades of years might be required for an initiated cell to become a fully-pledged, metastasized tumor. DNA mutations are accumulated during this process including background mutations that emerge scholastically, as well as driver mutations that selectively occur in a handful of cancer genes and confer the cell a growth advantage over its neighbors. A clone of tumor cells could be superseded by another clone that acquires new mutations and grows more aggressively. Tumor evolutional patterns have been studied for years using conventional approaches that focus on the investigation of a single or a couple of genes. Latest deep sequencing technology enables a global view of tumor evolution by deciphering almost all genome aberrations in a tumor. Tumor clones and the fate of each clone during tumor evolution can be depicted with the help of the concept of variant allele frequency. Here, we summarize the new insights of cancer evolutional progression in acute myeloid leukemia. Cancer evolution is currently thought to start from a clone that has accumulated the requisite somatically-acquired genetic aberrations through a series of increasingly disordered clinical and pathological phases, eventually leading to malignant transformation [1-3]. The observations in invasive colorectal cancer that usually emerges from an antecedent benign adenomatous polyp and in cervical cancer that proceeds through intraepithelial neoplasia support the idea of stepwise or linear cancerous progression [3-5]. Genetically, such progression is achieved by successive waves of clonal expansion during which cells acquire novel genomic alterations including single nucleotide variants (SNVs), small insertions and deletions (indels), and/or copy number variations (CNVs) [6]. The latest improvement in sequencing technology has allowed the deciphering of the whole exome or genome in different types of tumor and normal tissue pairs, providing detailed catalogue about genome aberrations during tumor initiation and progression, which have been reviewed in several papers [7-10]. Here, we focus on demonstrating the cancer clonal evolution pattern revealed by recent deep sequencing studies of samples from acute myeloid leukemia (AML) patients.
Keywords: acute myeloid leukemia; cancer genome; clonal evolution.
Conflict of interest statement
We do not have any conflicts of interest.
Figures
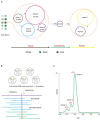
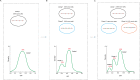


Similar articles
-
Genomic complexity and dynamics of clonal evolution in childhood acute myeloid leukemia studied with whole-exome sequencing.Oncotarget. 2016 Aug 30;7(35):56746-56757. doi: 10.18632/oncotarget.10778. Oncotarget. 2016. PMID: 27462774 Free PMC article.
-
Close correlation of copy number aberrations detected by next-generation sequencing with results from routine cytogenetics in acute myeloid leukemia.Genes Chromosomes Cancer. 2016 Jul;55(7):553-67. doi: 10.1002/gcc.22359. Epub 2016 May 2. Genes Chromosomes Cancer. 2016. PMID: 27015608
-
Whole-exome sequencing reveals the spectrum of gene mutations and the clonal evolution patterns in paediatric acute myeloid leukaemia.Br J Haematol. 2016 Nov;175(3):476-489. doi: 10.1111/bjh.14247. Epub 2016 Jul 29. Br J Haematol. 2016. PMID: 27470916
-
Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia.Leukemia. 2015 Jun;29(6):1243-52. doi: 10.1038/leu.2015.32. Epub 2015 Feb 12. Leukemia. 2015. PMID: 25673237 Review.
-
Detection of copy number alterations in acute myeloid leukemia and myelodysplastic syndromes.Expert Rev Mol Diagn. 2012 Apr;12(3):253-64. doi: 10.1586/erm.12.18. Expert Rev Mol Diagn. 2012. PMID: 22468816 Review.
Cited by
-
Endothelial cells are a key target of IFN-g during response to combined PD-1/CTLA-4 ICB treatment in a mouse model of bladder cancer.iScience. 2023 Sep 15;26(10):107937. doi: 10.1016/j.isci.2023.107937. eCollection 2023 Oct 20. iScience. 2023. PMID: 37810214 Free PMC article.
-
Practical Considerations for Treatment of Relapsed/Refractory FLT3-ITD Acute Myeloid Leukaemia with Quizartinib: Illustrative Case Reports.Clin Drug Investig. 2020 Mar;40(3):227-235. doi: 10.1007/s40261-019-00881-7. Clin Drug Investig. 2020. PMID: 31912423 Free PMC article. Review.
-
Colony Stimulating Factor 1 Receptor in Acute Myeloid Leukemia.Front Oncol. 2021 Mar 25;11:654817. doi: 10.3389/fonc.2021.654817. eCollection 2021. Front Oncol. 2021. PMID: 33842370 Free PMC article. Review.
-
From Samples to Germline and Somatic Sequence Variation: A Focus on Next-Generation Sequencing in Melanoma Research.Life (Basel). 2022 Nov 21;12(11):1939. doi: 10.3390/life12111939. Life (Basel). 2022. PMID: 36431075 Free PMC article. Review.
-
Co-evolution of tumor and immune cells during progression of multiple myeloma.Nat Commun. 2021 May 7;12(1):2559. doi: 10.1038/s41467-021-22804-x. Nat Commun. 2021. PMID: 33963182 Free PMC article.
References
-
- Castro-Giner F, Ratcliffe P, Tomlinson I. The mini-driver model of polygenic cancer evolution. Nature reviews Cancer. 2015;15:680–685. - PubMed
-
- Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nature reviews Cancer. 2015;15:473–483. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Farber E. The multistep nature of cancer development. Cancer research. 1984;44:4217–4223. - PubMed
-
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends in genetics. 1993;9:138–141. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

