A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation
- PMID: 27474443
- PMCID: PMC4973295
- DOI: 10.1101/gad.284604.116
A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation
Abstract
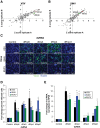
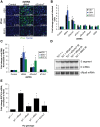
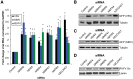
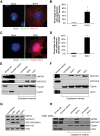
RNA degradation is tightly regulated to selectively target aberrant RNAs, including viral RNA, but this regulation is incompletely understood. Through RNAi screening in Drosophila cells, we identified the 3'-to-5' RNA exosome and two components of the exosome cofactor TRAMP (Trf4/5-Air1/2-Mtr4 polyadenylation) complex, dMtr4 and dZcchc7, as antiviral against a panel of RNA viruses. We extended our studies to human orthologs and found that the exosome as well as TRAMP components hMTR4 and hZCCHC7 are antiviral. While hMTR4 and hZCCHC7 are normally nuclear, infection by cytoplasmic RNA viruses induces their export, forming a cytoplasmic complex that specifically recognizes and induces degradation of viral mRNAs. Furthermore, the 3' untranslated region (UTR) of bunyaviral mRNA is sufficient to confer virus-induced exosomal degradation. Altogether, our results reveal that signals from viral infection repurpose TRAMP components to a cytoplasmic surveillance role where they selectively engage viral RNAs for degradation to restrict a broad range of viruses.
Keywords: RNA degradation; TRAMP; antiviral; arbovirus; exosome; intrinsic immunity.
© 2016 Molleston et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex.J Biol Chem. 2011 Oct 28;286(43):37429-45. doi: 10.1074/jbc.M111.271494. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878619 Free PMC article.
-
Purification of Endogenous Tagged TRAMP4/5 and Exosome Complexes from Yeast and In Vitro Polyadenylation-Exosome Activation Assays.Methods Mol Biol. 2020;2062:237-253. doi: 10.1007/978-1-4939-9822-7_12. Methods Mol Biol. 2020. PMID: 31768980
-
TRAMP Stimulation of Exosome.Enzymes. 2012;31:77-95. doi: 10.1016/B978-0-12-404740-2.00004-5. Epub 2012 Sep 29. Enzymes. 2012. PMID: 27166441
-
Comparison of the yeast and human nuclear exosome complexes.Biochem Soc Trans. 2012 Aug;40(4):850-5. doi: 10.1042/BST20120061. Biochem Soc Trans. 2012. PMID: 22817747 Review.
-
Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity.Wiley Interdiscip Rev RNA. 2013 Mar-Apr;4(2):217-31. doi: 10.1002/wrna.1155. Epub 2013 Feb 15. Wiley Interdiscip Rev RNA. 2013. PMID: 23417976 Free PMC article. Review.
Cited by
-
RNA helicase SKIV2L limits antiviral defense and autoinflammation elicited by the OAS-RNase L pathway.EMBO J. 2024 Sep;43(18):3876-3894. doi: 10.1038/s44318-024-00187-1. Epub 2024 Aug 7. EMBO J. 2024. PMID: 39112803 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus ORF57 protein protects viral transcripts from specific nuclear RNA decay pathways by preventing hMTR4 recruitment.PLoS Pathog. 2019 Feb 20;15(2):e1007596. doi: 10.1371/journal.ppat.1007596. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30785952 Free PMC article.
-
ZCCHC3 is a stress granule zinc knuckle protein that strongly suppresses LINE-1 retrotransposition.PLoS Genet. 2023 Jul 5;19(7):e1010795. doi: 10.1371/journal.pgen.1010795. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37405998 Free PMC article.
-
Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors.Genes Dev. 2017 Jan 15;31(2):88-100. doi: 10.1101/gad.294769.116. Genes Dev. 2017. PMID: 28202538 Free PMC article. Review.
-
Attacked from All Sides: RNA Decay in Antiviral Defense.Viruses. 2017 Jan 4;9(1):2. doi: 10.3390/v9010002. Viruses. 2017. PMID: 28054965 Free PMC article. Review.
References
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. 2011. Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29: 185–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
