Development and validation of a cell-based assay system to assess human immunodeficiency virus type 1 integrase multimerization
- PMID: 27474494
- PMCID: PMC8188399
- DOI: 10.1016/j.jviromet.2016.07.023
Development and validation of a cell-based assay system to assess human immunodeficiency virus type 1 integrase multimerization
Abstract
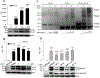
Multimerization of HIV-1 integrase (IN) subunits is required for the concerted integration of HIV-1 proviral DNA into the host genome. Thus, the disruption of IN multimerization represents a new avenue for intervening HIV-1 infection. Here, we generated a cell-based assay system to assess IN multimerization using a newly constructed bimolecular fluorescence complementation (BiFC-IN) system. BiFC-IN proteins were efficient in emitting fluorescence, and amino acid (AA) substitutions associated with IN multimerization attenuated fluorescence, suggesting that the BiFC-IN system may be useful for evaluating the profile of IN multimerization. A recently reported non-catalytic site IN inhibitor (NCINI), which allosterically induces IN over-multimerization/aggregation, significantly increased fluorescence in the BiFC-IN system. An IN's substitution, A128T, associated with viral resistance to NCINIs, decreased the NCINI-induced increase of fluorescence, suggesting that A128T reduces the potential for IN over-multimerization. Moreover, E11K and F181T substitutions known to inhibit IN tetramerization also reduced the NCINI-induced fluorescence increase, suggesting that NCINI-induced IN over-multimerization was more likely to occur from tetramer subunits than from dimer subunits. The present study demonstrates that our cell-based BiFC-IN system may be useful in elucidating the profile of IN multimerization, and also help evaluate and identify novel compounds that disrupt IN multimerization in living cells.
Keywords: BiFC; HIV-1; HIV-1 integrase multimerization; NCINIs.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


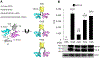


References
-
- Al-Mawsawi LQ, Hombrouck A, Dayam R, Debyser Z, Neamati N, 2008. Four-tiered π interaction at the dimeric interface of HIV-1 integrase critical for DNA integration and viral infectivity. Virology 377, 355–363. - PubMed
-
- Amano M, Koh Y, Das D, Li J, Leschenko S, Wang YF, Boross PI, Weber IT, Ghosh AK, Mitsuya H, 2007. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother 51, 2143–2155. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

