Transcriptional Control in Marine Copiotrophic and Oligotrophic Bacteria with Streamlined Genomes
- PMID: 27474718
- PMCID: PMC5038029
- DOI: 10.1128/AEM.01299-16
Transcriptional Control in Marine Copiotrophic and Oligotrophic Bacteria with Streamlined Genomes
Abstract
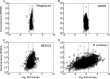
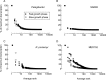
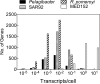
Bacteria often respond to environmental stimuli using transcriptional control, but this may not be the case for marine bacteria such as "Candidatus Pelagibacter ubique," a cultivated representative of the SAR11 clade, the most abundant organism in the ocean. This bacterium has a small, streamlined genome and an unusually low number of transcriptional regulators, suggesting that transcriptional control is low in Pelagibacter and limits its response to environmental conditions. Transcriptome sequencing during batch culture growth revealed that only 0.1% of protein-encoding genes appear to be under transcriptional control in Pelagibacter and in another oligotroph (SAR92) whereas >10% of genes were under transcriptional control in the copiotrophs Polaribacter sp. strain MED152 and Ruegeria pomeroyi When growth levels changed, transcript levels remained steady in Pelagibacter and SAR92 but shifted in MED152 and R. pomeroyi Transcript abundances per cell, determined using an internal RNA sequencing standard, were low (<1 transcript per cell) for all but a few of the most highly transcribed genes in all four taxa, and there was no correlation between transcript abundances per cell and shifts in the levels of transcription. These results suggest that low transcriptional control contributes to the success of Pelagibacter and possibly other oligotrophic microbes that dominate microbial communities in the oceans.
Importance: Diverse heterotrophic bacteria drive biogeochemical cycling in the ocean. The most abundant types of marine bacteria are oligotrophs with small, streamlined genomes. The metabolic controls that regulate the response of oligotrophic bacteria to environmental conditions remain unclear. Our results reveal that transcriptional control is lower in marine oligotrophic bacteria than in marine copiotrophic bacteria. Although responses of bacteria to environmental conditions are commonly regulated at the level of transcription, metabolism in the most abundant bacteria in the ocean appears to be regulated by other mechanisms.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Wear EK, Carlson CA, James AK, Brzezinski MA, Windecker LA, Nelson CE. 2015. Synchronous shifts in dissolved organic carbon bioavailability and bacterial community responses over the course of an upwelling-driven phytoplankton bloom. Limnol Oceanogr 60:657–677. doi:10.1002/lno.10042. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

