A network based approach to drug repositioning identifies plausible candidates for breast cancer and prostate cancer
- PMID: 27475327
- PMCID: PMC4967295
- DOI: 10.1186/s12920-016-0212-7
A network based approach to drug repositioning identifies plausible candidates for breast cancer and prostate cancer
Abstract
Background: The high cost and the long time required to bring drugs into commerce is driving efforts to repurpose FDA approved drugs-to find new uses for which they weren't intended, and to thereby reduce the overall cost of commercialization, and shorten the lag between drug discovery and availability. We report on the development, testing and application of a promising new approach to repositioning.
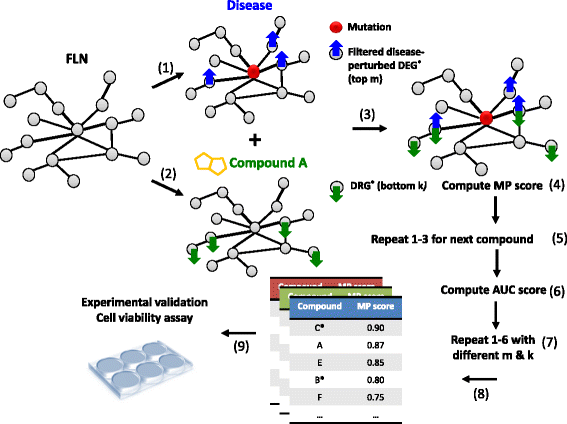
Methods: Our approach is based on mining a human functional linkage network for inversely correlated modules of drug and disease gene targets. The method takes account of multiple information sources, including gene mutation, gene expression, and functional connectivity and proximity of within module genes.
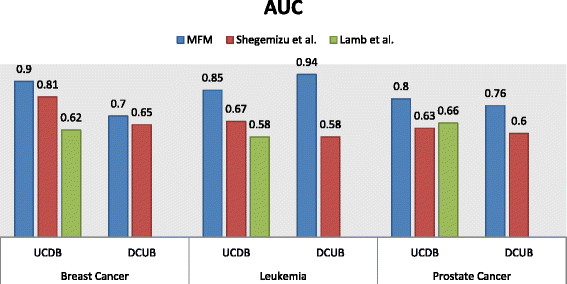
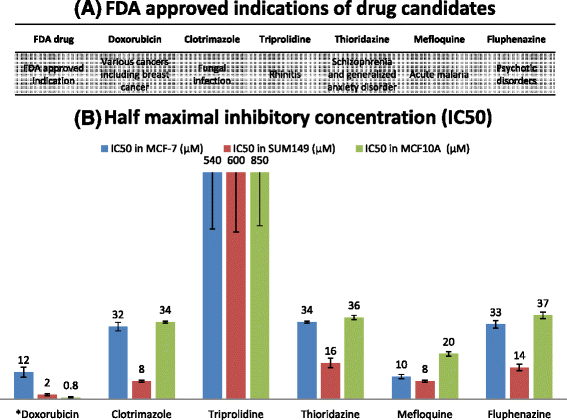
Results: The method was used to identify candidates for treating breast and prostate cancer. We found that (i) the recall rate for FDA approved drugs for breast (prostate) cancer is 20/20 (10/11), while the rates for drugs in clinical trials were 131/154 and 82/106; (ii) the ROC/AUC performance substantially exceeds that of comparable methods; (iii) preliminary in vitro studies indicate that 5/5 candidates have therapeutic indices superior to that of Doxorubicin in MCF7 and SUM149 cancer cell lines. We briefly discuss the biological plausibility of the candidates at a molecular level in the context of the biological processes that they mediate.
Conclusions: Our method appears to offer promise for the identification of multi-targeted drug candidates that can correct aberrant cellular functions. In particular the computational performance exceeded that of other CMap-based methods, and in vitro experiments indicate that 5/5 candidates have therapeutic indices superior to that of Doxorubicin in MCF7 and SUM149 cancer cell lines. The approach has the potential to provide a more efficient drug discovery pipeline.
Keywords: Cancer treatment; Computational drug repositioning; Drug screening.
Figures
Similar articles
-
Predicting Potential Drugs for Breast Cancer based on miRNA and Tissue Specificity.Int J Biol Sci. 2018 May 22;14(8):971-982. doi: 10.7150/ijbs.23350. eCollection 2018. Int J Biol Sci. 2018. PMID: 29989066 Free PMC article.
-
Drug repositioning in SLE: crowd-sourcing, literature-mining and Big Data analysis.Lupus. 2016 Sep;25(10):1150-70. doi: 10.1177/0961203316657437. Lupus. 2016. PMID: 27497259 Review.
-
Computational drugs repositioning identifies inhibitors of oncogenic PI3K/AKT/P70S6K-dependent pathways among FDA-approved compounds.Oncotarget. 2016 Sep 13;7(37):58743-58758. doi: 10.18632/oncotarget.11318. Oncotarget. 2016. PMID: 27542212 Free PMC article.
-
MD-Miner: a network-based approach for personalized drug repositioning.BMC Syst Biol. 2017 Oct 3;11(Suppl 5):86. doi: 10.1186/s12918-017-0462-9. BMC Syst Biol. 2017. PMID: 28984195 Free PMC article.
-
Drug repositioning: re-investigating existing drugs for new therapeutic indications.J Postgrad Med. 2011 Apr-Jun;57(2):153-60. doi: 10.4103/0022-3859.81870. J Postgrad Med. 2011. PMID: 21654146 Review.
Cited by
-
Novel therapeutic compounds for prostate adenocarcinoma treatment: An analysis using bioinformatic approaches and the CMap database.Medicine (Baltimore). 2020 Dec 18;99(51):e23768. doi: 10.1097/MD.0000000000023768. Medicine (Baltimore). 2020. PMID: 33371142 Free PMC article.
-
In silico screening for ERα down modulators identifies thioridazine as an anti-proliferative agent in primary, 4OH-tamoxifen-resistant and Y537S ERα-expressing breast cancer cells.Cell Oncol (Dordr). 2018 Dec;41(6):677-686. doi: 10.1007/s13402-018-0400-x. Epub 2018 Sep 4. Cell Oncol (Dordr). 2018. PMID: 30182339
-
Network neighborhood operates as a drug repositioning method for cancer treatment.PeerJ. 2023 Jul 10;11:e15624. doi: 10.7717/peerj.15624. eCollection 2023. PeerJ. 2023. PMID: 37456868 Free PMC article.
-
The Discovery of New Drug-Target Interactions for Breast Cancer Treatment.Molecules. 2021 Dec 10;26(24):7474. doi: 10.3390/molecules26247474. Molecules. 2021. PMID: 34946556 Free PMC article.
-
Reconciling multiple connectivity-based systems biology methods for drug repurposing.Brief Bioinform. 2025 Jul 2;26(4):bbaf387. doi: 10.1093/bib/bbaf387. Brief Bioinform. 2025. PMID: 40736744 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

