Dynamic expression of the mouse orthologue of the human amyotropic lateral sclerosis associated gene C9orf72 during central nervous system development and neuronal differentiation
- PMID: 27476503
- PMCID: PMC5108160
- DOI: 10.1111/joa.12526
Dynamic expression of the mouse orthologue of the human amyotropic lateral sclerosis associated gene C9orf72 during central nervous system development and neuronal differentiation
Abstract
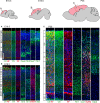
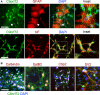
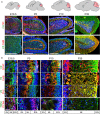
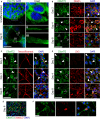
The hexanucleotide repeat in the first intron of the C9orf72 gene is the most significant cause of amyotropic lateral sclerosis as well as some forms of fronto-temporal dementia. The C9orf72 protein has been previously reported to be expressed in post-mortem human brain as well as in late embryonic and some postnatal stages in mice. Herein, we present a detailed study of the distribution of C9orf72 protein in the embryonic, postnatal and adult mouse brain, spinal cord as well as during the differentiation of P19 embryonal carcinoma cells to neurons including motor neurons. We show that the expression levels of the C9orf72 transcripts in the developing and adult mouse brain as well as in differentiating neurons, are dynamic. Besides the strong expression in the cerebellum and motor cortex reported previously, we show for the first time that C9orf72 is expressed strongly in the olfactory bulb and also in the hippocampus. Our immunostaining data also reveal a hitherto unreported switch in the cellular distribution of C9orf72 from a predominantly cytoplasmic to a nucleo-cytoplasmic distribution during corticogenesis. This switch in distribution was also observed during differentiation of the pluripotent embryonal carcinoma P19 cell line to mature neurons. Our findings have implications for interpreting the pathophysiology caused by the repeat expansions in C9orf72 in mouse models.
Keywords: C9orf72; brain development; expression; neuronal differentiation; spinal cord.
© 2016 Anatomical Society.
Figures






Similar articles
-
Expression of DA11, a neuronal-injury-induced fatty acid binding protein, coincides with axon growth and neuronal differentiation during central nervous system development.J Neurosci Res. 1997 Jun 15;48(6):551-62. doi: 10.1002/(sici)1097-4547(19970615)48:6<551::aid-jnr8>3.0.co;2-9. J Neurosci Res. 1997. PMID: 9210525
-
Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis.Ann Neurol. 2013 Aug;74(2):180-7. doi: 10.1002/ana.23946. Ann Neurol. 2013. PMID: 23720273
-
Immunohistochemical detection of C9orf72 protein in frontotemporal lobar degeneration and motor neurone disease: patterns of immunostaining and an evaluation of commercial antibodies.Amyotroph Lateral Scler Frontotemporal Degener. 2018 Feb;19(1-2):102-111. doi: 10.1080/21678421.2017.1359304. Epub 2017 Aug 2. Amyotroph Lateral Scler Frontotemporal Degener. 2018. PMID: 28766957 Free PMC article.
-
Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review.
-
C9orf72 isoforms in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration.Brain Res. 2016 Sep 15;1647:43-49. doi: 10.1016/j.brainres.2016.04.062. Epub 2016 Apr 29. Brain Res. 2016. PMID: 27134035 Review.
Cited by
-
Cell-type specific differences in promoter activity of the ALS-linked C9orf72 mouse ortholog.Sci Rep. 2017 Jul 18;7(1):5685. doi: 10.1038/s41598-017-05864-2. Sci Rep. 2017. PMID: 28720882 Free PMC article.
-
C9orf72-mediated ALS and FTD: multiple pathways to disease.Nat Rev Neurol. 2018 Sep;14(9):544-558. doi: 10.1038/s41582-018-0047-2. Nat Rev Neurol. 2018. PMID: 30120348 Free PMC article. Review.
-
Molecular pathology, developmental changes and synaptic dysfunction in (pre-) symptomatic human C9ORF72-ALS/FTD cerebral organoids.Acta Neuropathol Commun. 2024 Sep 18;12(1):152. doi: 10.1186/s40478-024-01857-1. Acta Neuropathol Commun. 2024. PMID: 39289761 Free PMC article.
-
C9ORF72: What It Is, What It Does, and Why It Matters.Front Cell Neurosci. 2021 May 5;15:661447. doi: 10.3389/fncel.2021.661447. eCollection 2021. Front Cell Neurosci. 2021. PMID: 34025358 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

