Circadian Rhythm Disruption Promotes Lung Tumorigenesis
- PMID: 27476975
- PMCID: PMC5367626
- DOI: 10.1016/j.cmet.2016.07.001
Circadian Rhythm Disruption Promotes Lung Tumorigenesis
Abstract
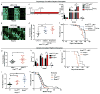
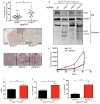
Circadian rhythms are 24-hr oscillations that control a variety of biological processes in living systems, including two hallmarks of cancer, cell division and metabolism. Circadian rhythm disruption by shift work is associated with greater risk for cancer development and poor prognosis, suggesting a putative tumor-suppressive role for circadian rhythm homeostasis. Using a genetically engineered mouse model of lung adenocarcinoma, we have characterized the effects of circadian rhythm disruption on lung tumorigenesis. We demonstrate that both physiologic perturbation (jet lag) and genetic mutation of the central circadian clock components decreased survival and promoted lung tumor growth and progression. The core circadian genes Per2 and Bmal1 were shown to have cell-autonomous tumor-suppressive roles in transformation and lung tumor progression. Loss of the central clock components led to increased c-Myc expression, enhanced proliferation, and metabolic dysregulation. Our findings demonstrate that both systemic and somatic disruption of circadian rhythms contribute to cancer progression.
Keywords: CRISPR; circadian rhythms; jet lag; lung cancer; physiology.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Alhopuro P, Bjorklund M, Sammalkorpi H, Turunen M, Tuupanen S, Bistrom M, Niittymaki I, Lehtonen HJ, Kivioja T, Launonen V, et al. Mutations in the circadian gene CLOCK in colorectal cancer. Mol Cancer Res. 2010;8:952–960. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

