Nucleases from Prevotella intermedia can degrade neutrophil extracellular traps
- PMID: 27476978
- PMCID: PMC5516193
- DOI: 10.1111/omi.12171
Nucleases from Prevotella intermedia can degrade neutrophil extracellular traps
Abstract
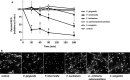
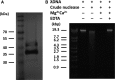
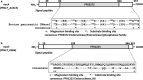
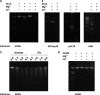
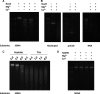
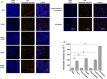
Periodontitis is an inflammatory disease caused by periodontal bacteria in subgingival plaque. These bacteria are able to colonize the periodontal region by evading the host immune response. Neutrophils, the host's first line of defense against infection, use various strategies to kill invading pathogens, including neutrophil extracellular traps (NETs). These are extracellular net-like fibers comprising DNA and antimicrobial components such as histones, LL-37, defensins, myeloperoxidase, and neutrophil elastase from neutrophils that disarm and kill bacteria extracellularly. Bacterial nuclease degrades the NETs to escape NET killing. It has now been shown that extracellular nucleases enable bacteria to evade this host antimicrobial mechanism, leading to increased pathogenicity. Here, we compared the DNA degradation activity of major Gram-negative periodontopathogenic bacteria, Porphyromonas gingivalis, Prevotella intermedia, Fusobacterium nucleatum, and Aggregatibacter actinomycetemcomitans. We found that Pr. intermedia showed the highest DNA degradation activity. A genome search of Pr. intermedia revealed the presence of two genes, nucA and nucD, putatively encoding secreted nucleases, although their enzymatic and biological activities are unknown. We cloned nucA- and nucD-encoding nucleases from Pr. intermedia ATCC 25611 and characterized their gene products. Recombinant NucA and NucD digested DNA and RNA, which required both Mg2+ and Ca2+ for optimal activity. In addition, NucA and NucD were able to degrade the DNA matrix comprising NETs.
Keywords: nucA; nucD; deoxyribonucleas; neutrophil extracellular trap degradation; periodontal disease.
© 2016 The Authors Molecular Oral Microbiology Published by John Wiley & Sons Ltd.
Figures

References
-
- Beiter, K. , Wartha, F. , Albiger, B. , Normark, S. , Zychlinsky, A. and Henriques‐Normark, B. (2006) An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol 16: 401–407. - PubMed
-
- Brinkmann, V. , Reichard, U. , Goosmann, C. et al (2004) Neutrophil extracellular traps kill bacteria. Science 303: 1532–1535. - PubMed
-
- Buchanan, J.T. , Simpson, A.J. , Aziz, R.K. et al (2006) DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol 16: 396–400. - PubMed
-
- de Buhr, N. , Neumann, A. , Jerjomiceva, N. , von Kockritz‐Blickwede, M. and Baums, C.G. (2014) Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET‐mediated antimicrobial activity. Microbiology 160: 385–395. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

