A Transcript-Specific eIF3 Complex Mediates Global Translational Control of Energy Metabolism
- PMID: 27477275
- PMCID: PMC4988921
- DOI: 10.1016/j.celrep.2016.07.006
A Transcript-Specific eIF3 Complex Mediates Global Translational Control of Energy Metabolism
Abstract
The multi-subunit eukaryotic translation initiation factor eIF3 is thought to assist in the recruitment of ribosomes to mRNA. The expression of eIF3 subunits is frequently disrupted in human cancers, but the specific roles of individual subunits in mRNA translation and cancer remain elusive. Using global transcriptomic, proteomic, and metabolomic profiling, we found a striking failure of Schizosaccharomyces pombe cells lacking eIF3e and eIF3d to synthesize components of the mitochondrial electron transport chain, leading to a defect in respiration, endogenous oxidative stress, and premature aging. Energy balance was maintained, however, by a switch to glycolysis with increased glucose uptake, upregulation of glycolytic enzymes, and strict dependence on a fermentable carbon source. This metabolic regulatory function appears to be conserved in human cells where eIF3e binds metabolic mRNAs and promotes their translation. Thus, via its eIF3d-eIF3e module, eIF3 orchestrates an mRNA-specific translational mechanism controlling energy metabolism that may be disrupted in cancer.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


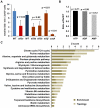
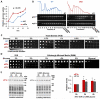


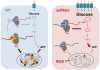
Comment in
-
Translation Factors Specify Cellular Metabolic State.Cell Rep. 2016 Aug 16;16(7):1787-8. doi: 10.1016/j.celrep.2016.07.067. Cell Rep. 2016. PMID: 27533178
Similar articles
-
Fission yeast translation initiation factor 3 subunit eIF3h is not essential for global translation initiation, but deletion of eif3h+ affects spore formation.Yeast. 2008 Nov;25(11):809-23. doi: 10.1002/yea.1635. Yeast. 2008. PMID: 19061185
-
Translational activators and mitoribosomal isoforms cooperate to mediate mRNA-specific translation in Schizosaccharomyces pombe mitochondria.Nucleic Acids Res. 2021 Nov 8;49(19):11145-11166. doi: 10.1093/nar/gkab789. Nucleic Acids Res. 2021. PMID: 34634819 Free PMC article.
-
Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle.Nucleic Acids Res. 2017 Nov 2;45(19):10948-10968. doi: 10.1093/nar/gkx805. Nucleic Acids Res. 2017. PMID: 28981723 Free PMC article.
-
The role of eIF3 and its individual subunits in cancer.Biochim Biophys Acta. 2015 Jul;1849(7):792-800. doi: 10.1016/j.bbagrm.2014.10.005. Epub 2014 Nov 1. Biochim Biophys Acta. 2015. PMID: 25450521 Review.
-
Human eIF3: from 'blobology' to biological insight.Philos Trans R Soc Lond B Biol Sci. 2017 Mar 19;372(1716):20160176. doi: 10.1098/rstb.2016.0176. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28138064 Free PMC article. Review.
Cited by
-
eIF3 interacts with histone H4 messenger RNA to regulate its translation.J Biol Chem. 2021 Jan-Jun;296:100578. doi: 10.1016/j.jbc.2021.100578. Epub 2021 Mar 23. J Biol Chem. 2021. PMID: 33766559 Free PMC article.
-
eIF-Three to Tango: emerging functions of translation initiation factor eIF3 in protein synthesis and disease.J Mol Cell Biol. 2020 Jul 3;12(6):403-409. doi: 10.1093/jmcb/mjaa018. J Mol Cell Biol. 2020. PMID: 32279082 Free PMC article. Review.
-
Functional and molecular profiling of fasted piglets reveals decreased energy metabolic function and cell proliferation in the small intestine.Am J Physiol Gastrointest Liver Physiol. 2023 Dec 1;325(6):G539-G555. doi: 10.1152/ajpgi.00240.2022. Epub 2023 Oct 17. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 37847725 Free PMC article.
-
Protein synthesis control in cancer: selectivity and therapeutic targeting.EMBO J. 2022 Apr 19;41(8):e109823. doi: 10.15252/embj.2021109823. Epub 2022 Mar 22. EMBO J. 2022. PMID: 35315941 Free PMC article. Review.
-
The Cap-Binding Complex CBC and the Eukaryotic Translation Factor eIF4E: Co-Conspirators in Cap-Dependent RNA Maturation and Translation.Cancers (Basel). 2021 Dec 8;13(24):6185. doi: 10.3390/cancers13246185. Cancers (Basel). 2021. PMID: 34944805 Free PMC article. Review.
References
-
- Asano K, Merrick WC, Hershey JWB. The Translation Initiation Factor eIF3-p48 Subunit Is Encoded by int-6, a Site of Frequent Integration by the Mouse Mammary Tumor Virus Genome. J. Biol. Chem. 1997;272:23477–23480. - PubMed
-
- Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the Fission Yeast Homologues of Mammalian Translation Initiation Factor 3 Subunits p66 (eIF3d) and p48 (eIF3e), Respectively, Are Required for Stable Association of eIF3 Subunits. J. Biol. Chem. 2002;277:2360–2367. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

