ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
- PMID: 27477906
- PMCID: PMC4975651
- DOI: 10.1016/j.molcel.2016.06.035
ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Abstract
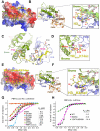
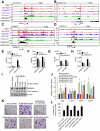
Histone acetylation, including acetylated H3K14 (H3K14ac), is generally linked to gene activation. Monomethylated histone H3 lysine 4 (H3K4me1), together with other gene-activating marks, denotes active genes. In contrast to usual gene-activating functions of H3K14ac and H3K4me1, we here show that the dual histone modification mark H3K4me1-H3K14ac is recognized by ZMYND8 (also called RACK7) and can function to counteract gene expression. We identified ZMYND8 as a transcriptional corepressor of the H3K4 demethylase JARID1D. ZMYND8 antagonized the expression of metastasis-linked genes, and its knockdown increased the cellular invasiveness in vitro and in vivo. The plant homeodomain (PHD) and Bromodomain cassette in ZMYND8 mediated the combinatorial recognition of H3K4me1-H3K14ac and H3K4me0-H3K14ac by ZMYND8. These findings uncover an unexpected role for the signature H3K4me1-H3K14ac in attenuating gene expression and reveal a metastasis-suppressive epigenetic mechanism in which ZMYND8's PHD-Bromo cassette couples H3K4me1-H3K14ac with downregulation of metastasis-linked genes.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
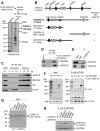
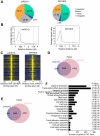
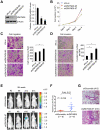
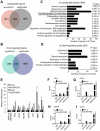

References
-
- Adhikary S, Sanyal S, Basu M, Sengupta I, Sen S, Srivastava DK, Roy S, Das C. Selective Recognition of H3.1K36 Dimethylation/H4K16 Acetylation Facilitates the Regulation of All-trans-retinoic Acid (ATRA)-responsive Genes by Putative Chromatin Reader ZMYND8. The Journal of biological chemistry. 2016;291:2664–2681. - PMC - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

