PADI2 gene confers susceptibility to breast cancer and plays tumorigenic role via ACSL4, BINC3 and CA9 signaling
- PMID: 27478411
- PMCID: PMC4966586
- DOI: 10.1186/s12935-016-0335-0
PADI2 gene confers susceptibility to breast cancer and plays tumorigenic role via ACSL4, BINC3 and CA9 signaling
Abstract
Background: Peptidylarginine deiminase (PAD) post-translationally converts arginine residues to citrulline residues. Recent studies have suggested that PADI2 (PAD isoform 2), a member of the PAD family, is involved in the tumorigenic process of some tumors, especially breast cancer. However, little is known about the mechanisms of PADI2 in tumorigenesis. This study aimed to elucidate the tumorigenic role and regulatory pathway of PADI2 in breast tumors.
Methods: The Sequenom MassARRAY and TaqMan genotyping methods were used to investigate the correlation between PADI2 gene SNPs and various tumor risks. PCR array analyses, including cancer pathway finder and signal transduction PCR arrays, were performed to investigate the tumorigenic pathway of PADI2 in the MCF-7 breast cancer cell line following treatment with anti-PADI2 siRNA. Cell proliferation, apoptosis and transwell migration assays were performed to observe the effect of PADI2 in MCF-7 cells treated with anti-PADI2 siRNA.
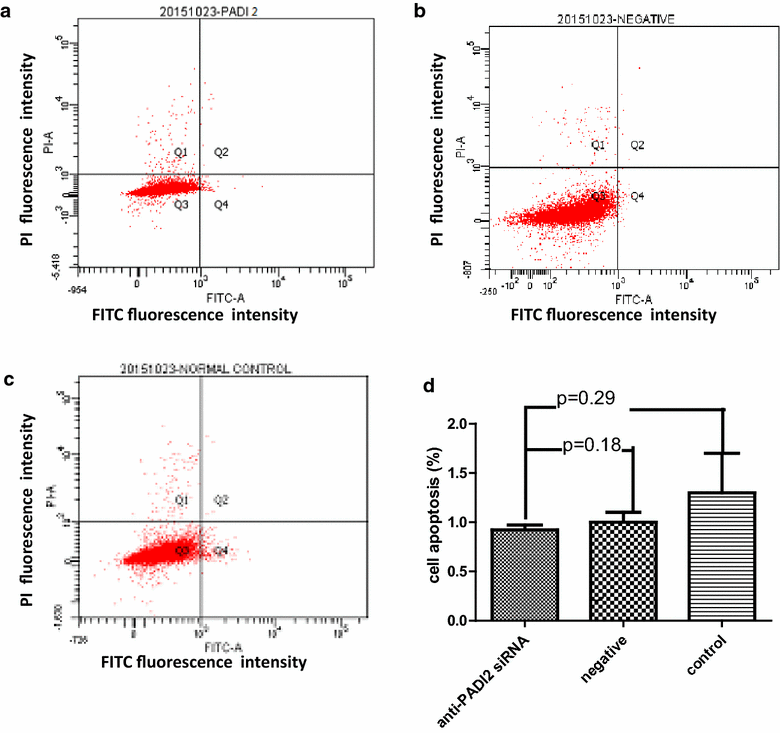
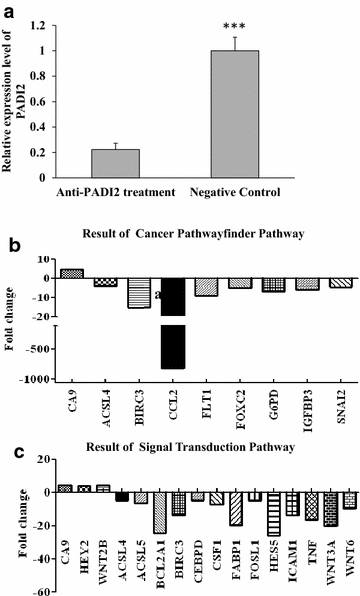
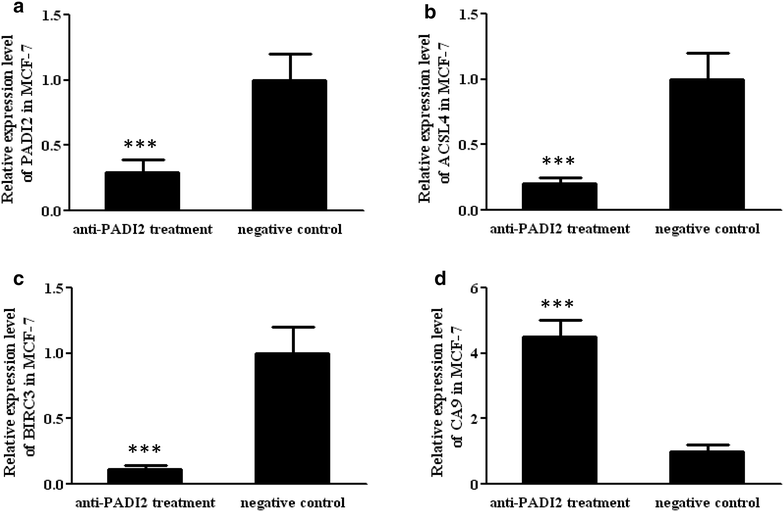
Results: Both Sequenom MassARRAY and TaqMan genotyping assays demonstrated that SNP rs10788656 in the PADI2 gene was significantly associated with breast cancer. PCR arrays indicated that inhibiting PADI2 expression significantly increased expression of CA9 and decreased expression of ACSL4 and BIRC3 in MCF-7 cells, which was verified using real-time PCR. Inhibiting PADI2 expression also significantly decreased the migration ability of MCF-7 cells but did not affect cell proliferation or apoptosis.
Conclusions: The PADI2 gene confers susceptibility to breast cancer. PADI2 expression contributes to abnormal migration of breast tumor cells. PADI2 affects tumorigenesis in breast tumor cells by regulating the expression of ACSL4, BINC3 and CA9, which are known to promote abnormal lipid metabolism and cell invasion of tumors.
Keywords: ACSL4 (long-chain fatty acyl-CoA synthetase 4); BIRC3 (baculoviral IAP repeat containing 3); CA9 (carbonic anhydrase IX); Citrullination; PADI2 (peptidylarginine deiminase isoform 2); Peptidylarginine deiminase (PAD).
Figures


Similar articles
-
Investigating the expression, effect and tumorigenic pathway of PADI2 in tumors.Onco Targets Ther. 2017 Mar 8;10:1475-1485. doi: 10.2147/OTT.S92389. eCollection 2017. Onco Targets Ther. 2017. PMID: 28331341 Free PMC article.
-
PADI2 is significantly associated with rheumatoid arthritis.PLoS One. 2013 Dec 5;8(12):e81259. doi: 10.1371/journal.pone.0081259. eCollection 2013. PLoS One. 2013. PMID: 24339914 Free PMC article.
-
Identification of PADI2 as a potential breast cancer biomarker and therapeutic target.BMC Cancer. 2012 Oct 30;12:500. doi: 10.1186/1471-2407-12-500. BMC Cancer. 2012. PMID: 23110523 Free PMC article.
-
ACSL4 as a Potential Target and Biomarker for Anticancer: From Molecular Mechanisms to Clinical Therapeutics.Front Pharmacol. 2022 Jul 13;13:949863. doi: 10.3389/fphar.2022.949863. eCollection 2022. Front Pharmacol. 2022. PMID: 35910359 Free PMC article. Review.
-
Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism.Prostaglandins Other Lipid Mediat. 2019 Oct;144:106363. doi: 10.1016/j.prostaglandins.2019.106363. Epub 2019 Jul 12. Prostaglandins Other Lipid Mediat. 2019. PMID: 31306767 Review.
Cited by
-
High-throughput proteomics of breast cancer interstitial fluid: identification of tumor subtype-specific serologically relevant biomarkers.Mol Oncol. 2021 Feb;15(2):429-461. doi: 10.1002/1878-0261.12850. Epub 2021 Jan 4. Mol Oncol. 2021. PMID: 33176066 Free PMC article. Clinical Trial.
-
The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer.Oncotarget. 2017 Aug 3;8(41):70642-70652. doi: 10.18632/oncotarget.19956. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050308 Free PMC article.
-
Emerging Mechanisms and Treatment Progress on Liver Metastasis of Colorectal Cancer.Onco Targets Ther. 2021 May 6;14:3013-3036. doi: 10.2147/OTT.S301371. eCollection 2021. Onco Targets Ther. 2021. PMID: 33986602 Free PMC article. Review.
-
Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells.J Exp Clin Cancer Res. 2019 Oct 10;38(1):414. doi: 10.1186/s13046-019-1404-8. J Exp Clin Cancer Res. 2019. PMID: 31601253 Free PMC article.
-
Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications.MedComm (2020). 2023 May 2;4(3):e261. doi: 10.1002/mco2.261. eCollection 2023 Jun. MedComm (2020). 2023. PMID: 37143582 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

