C13orf31 (FAMIN) is a central regulator of immunometabolic function
- PMID: 27478939
- PMCID: PMC6581540
- DOI: 10.1038/ni.3532
C13orf31 (FAMIN) is a central regulator of immunometabolic function
Abstract
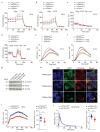
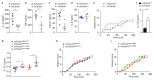
Single-nucleotide variations in C13orf31 (LACC1) that encode p.C284R and p.I254V in a protein of unknown function (called 'FAMIN' here) are associated with increased risk for systemic juvenile idiopathic arthritis, leprosy and Crohn's disease. Here we set out to identify the biological mechanism affected by these coding variations. FAMIN formed a complex with fatty acid synthase (FASN) on peroxisomes and promoted flux through de novo lipogenesis to concomitantly drive high levels of fatty-acid oxidation (FAO) and glycolysis and, consequently, ATP regeneration. FAMIN-dependent FAO controlled inflammasome activation, mitochondrial and NADPH-oxidase-dependent production of reactive oxygen species (ROS), and the bactericidal activity of macrophages. As p.I254V and p.C284R resulted in diminished function and loss of function, respectively, FAMIN determined resilience to endotoxin shock. Thus, we have identified a central regulator of the metabolic function and bioenergetic state of macrophages that is under evolutionary selection and determines the risk of inflammatory and infectious disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
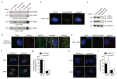
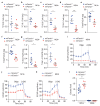

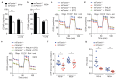
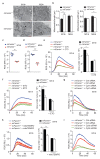

References
-
- Wakil SM, et al. Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis & rheumatology. 2015;67:288–295. - PubMed
-
- Patel N, et al. Study of Mendelian forms of Crohn's disease in Saudi Arabia reveals novel risk loci and alleles. Gut. 2014;63:1831–1832. - PubMed
-
- Liu H, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nature genetics. 2015;47:267–271. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 106260/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/000C0413/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 106260/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/000C0411/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U54 HG006348/HG/NHGRI NIH HHS/United States
- MC_UP_A090_1006/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/000C0417/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/P011705/1/MRC_/Medical Research Council/United Kingdom
- 079643/WT_/Wellcome Trust/United Kingdom
- 103077/WT_/Wellcome Trust/United Kingdom
- 260961/ERC_/European Research Council/International
- 100891/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/000C0415/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U42 OD011174/OD/NIH HHS/United States
- 100675/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

