Sensitive and Quantitative Three-Color Protein Imaging in Fission Yeast Using Spectrally Diverse, Recoded Fluorescent Proteins with Experimentally-Characterized In Vivo Maturation Kinetics
- PMID: 27479698
- PMCID: PMC4968791
- DOI: 10.1371/journal.pone.0159292
Sensitive and Quantitative Three-Color Protein Imaging in Fission Yeast Using Spectrally Diverse, Recoded Fluorescent Proteins with Experimentally-Characterized In Vivo Maturation Kinetics
Abstract
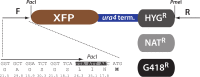
Schizosaccharomyces pombe is an outstanding model organism for cell biological investigations, yet the range of useful and well-characterized fluorescent proteins (XFPs) is limited. We generated and characterized three recoded fluorescent proteins for 3-color analysis in S.pombe, Super-folder GFP, monomeric Kusabira Orange 2 and E2Crimson. Upon optimization and expression in S. pombe, the three proteins enabled sensitive simultaneous 3-color detection capability. Furthermore, we describe a strategy that combines a pulse-chase approach and mathematical modeling to quantify the maturation kinetics of these proteins in vivo. We observed maturation kinetics in S. pombe that are expected from those described for these proteins in vitro and/or in other cell types, but also unpredicted behaviors. Our studies provide a kinetically-characterized, integrated three-color XFP toolbox for S. pombe.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

