A data set from flash X-ray imaging of carboxysomes
- PMID: 27479842
- PMCID: PMC4968195
- DOI: 10.1038/sdata.2016.61
A data set from flash X-ray imaging of carboxysomes
Abstract
Ultra-intense femtosecond X-ray pulses from X-ray lasers permit structural studies on single particles and biomolecules without crystals. We present a large data set on inherently heterogeneous, polyhedral carboxysome particles. Carboxysomes are cell organelles that vary in size and facilitate up to 40% of Earth's carbon fixation by cyanobacteria and certain proteobacteria. Variation in size hinders crystallization. Carboxysomes appear icosahedral in the electron microscope. A protein shell encapsulates a large number of Rubisco molecules in paracrystalline arrays inside the organelle. We used carboxysomes with a mean diameter of 115±26 nm from Halothiobacillus neapolitanus. A new aerosol sample-injector allowed us to record 70,000 low-noise diffraction patterns in 12 min. Every diffraction pattern is a unique structure measurement and high-throughput imaging allows sampling the space of structural variability. The different structures can be separated and phased directly from the diffraction data and open a way for accurate, high-throughput studies on structures and structural heterogeneity in biology and elsewhere.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


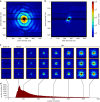
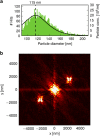

Comment in
-
The trickle before the torrent-diffraction data from X-ray lasers.Sci Data. 2016 Aug 1;3:160059. doi: 10.1038/sdata.2016.59. Sci Data. 2016. PMID: 27479637 Free PMC article.
References
Data Citations
-
- Hantke M. F. 2014. Coherent X-ray Imaging Data Bank. http://dx.doi.org/10.11577/1169545 - DOI
References
-
- Neutze R., Wouts R., van der Spoel D., Weckert E. & Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757 (2000). - PubMed
-
- Chapman H. N. et al. Femtosecond diffractive imaging with a soft-X-ray free-electron laser. Nat. Phys 2, 839–843 (2006).
-
- Bergh M., Huldt G., Timneanu N., Maia F. R. N. C. & Hajdu J. Feasibility of imaging living cells at subnanometer resolutions by ultrafast X-ray diffraction. Q. Rev. Biophys. 41, 181–204 (2008). - PubMed
-
- Hantke M. F. et al. High-throughput imaging of heterogeneous cell organelles with an X-ray laser. Nat. Photonics 8, 943–949 (2014).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

