Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (Secale cereale L.)
- PMID: 27480157
- PMCID: PMC5069347
- DOI: 10.1007/s00122-016-2756-5
Model training across multiple breeding cycles significantly improves genomic prediction accuracy in rye (Secale cereale L.)
Abstract
Genomic prediction accuracy can be significantly increased by model calibration across multiple breeding cycles as long as selection cycles are connected by common ancestors. In hybrid rye breeding, application of genome-based prediction is expected to increase selection gain because of long selection cycles in population improvement and development of hybrid components. Essentially two prediction scenarios arise: (1) prediction of the genetic value of lines from the same breeding cycle in which model training is performed and (2) prediction of lines from subsequent cycles. It is the latter from which a reduction in cycle length and consequently the strongest impact on selection gain is expected. We empirically investigated genome-based prediction of grain yield, plant height and thousand kernel weight within and across four selection cycles of a hybrid rye breeding program. Prediction performance was assessed using genomic and pedigree-based best linear unbiased prediction (GBLUP and PBLUP). A total of 1040 S2 lines were genotyped with 16 k SNPs and each year testcrosses of 260 S2 lines were phenotyped in seven or eight locations. The performance gap between GBLUP and PBLUP increased significantly for all traits when model calibration was performed on aggregated data from several cycles. Prediction accuracies obtained from cross-validation were in the order of 0.70 for all traits when data from all cycles (N CS = 832) were used for model training and exceeded within-cycle accuracies in all cases. As long as selection cycles are connected by a sufficient number of common ancestors and prediction accuracy has not reached a plateau when increasing sample size, aggregating data from several preceding cycles is recommended for predicting genetic values in subsequent cycles despite decreasing relatedness over time.
Conflict of interest statement
The authors declare that they have no conflict of interest. Ethical standards The authors declare that the experiments comply with the current laws of Germany.
Figures

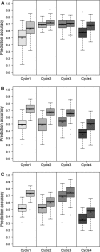
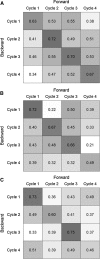
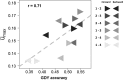
 ) prediction direction. Results are shown for all possible pairwise cycle combinations, with one cycle forming the calibration (N = 208) and one cycle the validation set (N = 52), respectively
) prediction direction. Results are shown for all possible pairwise cycle combinations, with one cycle forming the calibration (N = 208) and one cycle the validation set (N = 52), respectively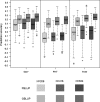
References
-
- Bauer E, Barilar I, Gundlach H, Hackauf B, Korzun V, Martis M, Mayer KFX, Schmid K, Schmutzer T, Schön C-C, Scholz U, Trost E (2015) Rye-don’t be afraid of an 8 Gb genome jigsaw. EUCARPIA-International Conference on Rye Breeding and Genetics, 24–26 June 2015, Wroclaw, Poland, pp 32–33
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

