Deep genome sequencing and variation analysis of 13 inbred mouse strains defines candidate phenotypic alleles, private variation and homozygous truncating mutations
- PMID: 27480531
- PMCID: PMC4968449
- DOI: 10.1186/s13059-016-1024-y
Deep genome sequencing and variation analysis of 13 inbred mouse strains defines candidate phenotypic alleles, private variation and homozygous truncating mutations
Abstract
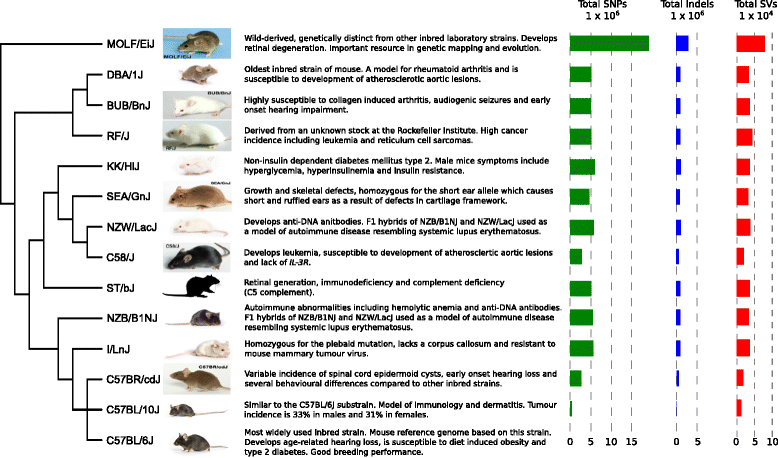
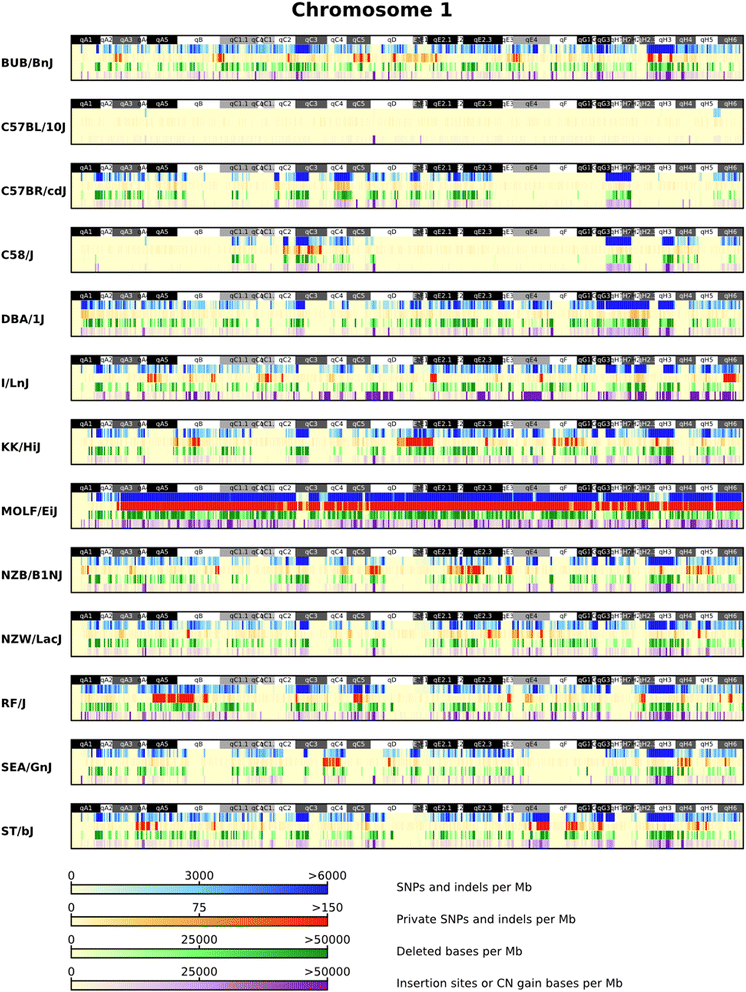
Background: The Mouse Genomes Project is an ongoing collaborative effort to sequence the genomes of the common laboratory mouse strains. In 2011, the initial analysis of sequence variation across 17 strains found 56.7 M unique single nucleotide polymorphisms (SNPs) and 8.8 M indels. We carry out deep sequencing of 13 additional inbred strains (BUB/BnJ, C57BL/10J, C57BR/cdJ, C58/J, DBA/1J, I/LnJ, KK/HiJ, MOLF/EiJ, NZB/B1NJ, NZW/LacJ, RF/J, SEA/GnJ and ST/bJ), cataloguing molecular variation within and across the strains. These strains include important models for immune response, leukaemia, age-related hearing loss and rheumatoid arthritis. We now have several examples of fully sequenced closely related strains that are divergent for several disease phenotypes.
Results: Approximately 27.4 M unique SNPs and 5 M indels are identified across these strains compared to the C57BL/6 J reference genome (GRCm38). The amount of variation found in the inbred laboratory mouse genome has increased to 71 M SNPs and 12 M indels. We investigate the genetic basis of highly penetrant cancer susceptibility in RF/J finding private novel missense mutations in DNA damage repair and highly cancer associated genes. We use two highly related strains (DBA/1J and DBA/2J) to investigate the genetic basis of collagen-induced arthritis susceptibility.
Conclusions: This paper significantly expands the catalogue of fully sequenced laboratory mouse strains and now contains several examples of highly genetically similar strains with divergent phenotypes. We show how studying private missense mutations can lead to insights into the genetic mechanism for a highly penetrant phenotype.
Keywords: Biological pathways; Cancer; Disease; Genomic variation; Laboratory mouse; Mouse genomes; Sequencing; arthritis.
Figures
Similar articles
-
Voluntary ethanol consumption in 22 inbred mouse strains.Alcohol. 2008 May;42(3):149-60. doi: 10.1016/j.alcohol.2007.12.006. Epub 2008 Mar 20. Alcohol. 2008. PMID: 18358676 Free PMC article.
-
Hearing loss associated with the modifier of deaf waddler (mdfw) locus corresponds with age-related hearing loss in 12 inbred strains of mice.Hear Res. 2001 Apr;154(1-2):45-53. doi: 10.1016/s0378-5955(01)00215-5. Hear Res. 2001. PMID: 11423214 Free PMC article.
-
Genomes of the Mouse Collaborative Cross.Genetics. 2017 Jun;206(2):537-556. doi: 10.1534/genetics.116.198838. Genetics. 2017. PMID: 28592495 Free PMC article.
-
Complete overview of protein-inactivating sequence variations in 36 sequenced mouse inbred strains.Proc Natl Acad Sci U S A. 2017 Aug 22;114(34):9158-9163. doi: 10.1073/pnas.1706168114. Epub 2017 Aug 7. Proc Natl Acad Sci U S A. 2017. PMID: 28784771 Free PMC article. Review.
-
PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies--a valuable resource of phenotypic variations and genomic polymorphisms.Folia Biol (Praha). 2000;46(1):31-41. Folia Biol (Praha). 2000. PMID: 10730880 Review.
Cited by
-
Variation under domestication in animal models: the case of the Mexican axolotl.BMC Genomics. 2020 Nov 23;21(1):827. doi: 10.1186/s12864-020-07248-9. BMC Genomics. 2020. PMID: 33228551 Free PMC article.
-
The potential of integrating human and mouse discovery platforms to advance our understanding of cardiometabolic diseases.Elife. 2023 Mar 31;12:e86139. doi: 10.7554/eLife.86139. Elife. 2023. PMID: 37000167 Free PMC article.
-
Whole Genome Sequence of Two Wild-Derived Mus musculus domesticus Inbred Strains, LEWES/EiJ and ZALENDE/EiJ, with Different Diploid Numbers.G3 (Bethesda). 2016 Dec 7;6(12):4211-4216. doi: 10.1534/g3.116.034751. G3 (Bethesda). 2016. PMID: 27765810 Free PMC article.
-
The genetic background significantly impacts the severity of kidney cystic disease in the Pkd1RC/RC mouse model of autosomal dominant polycystic kidney disease.Kidney Int. 2021 Jun;99(6):1392-1407. doi: 10.1016/j.kint.2021.01.028. Epub 2021 Mar 9. Kidney Int. 2021. PMID: 33705824 Free PMC article.
-
SNPs, short tandem repeats, and structural variants are responsible for differential gene expression across C57BL/6 and C57BL/10 substrains.Cell Genom. 2022 Mar 9;2(3):100102. doi: 10.1016/j.xgen.2022.100102. Cell Genom. 2022. PMID: 35720252 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

