Role of the Gut Microbiome in Modulating Arthritis Progression in Mice
- PMID: 27481047
- PMCID: PMC4969881
- DOI: 10.1038/srep30594
Role of the Gut Microbiome in Modulating Arthritis Progression in Mice
Abstract
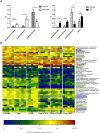
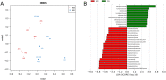
Genetics alone cannot explain most cases of rheumatoid arthritis (RA). Thus, investigating environmental factors such as the gut microbiota may provide new insights into the initiation and progression of RA. In this study, we performed 16S rRNA sequencing to characterise the gut microbiota of DBA1 mice that did or did not develop arthritis after induction with collagen. We found that divergence in the distribution of microbiota after induction was pronounced and significant. Mice susceptible to collagen-induced arthritis (CIA) showed enriched operational taxonomic units (OTUs) affiliated with the genus Lactobacillus as the dominant genus prior to arthritis onset. With disease development, the abundance of OTUs affiliated with the families Bacteroidaceae, Lachnospiraceae, and S24-7 increased significantly in CIA-susceptible mice. Notably, germ-free mice conventionalized with the microbiota from CIA-susceptible mice showed a higher frequency of arthritis induction than those conventionalized with the microbiota from CIA-resistant mice. Consistently, the concentration of the cytokine interleukin-17 in serum and the proportions of CD8+T cells and Th17 lymphocytes in the spleen were significantly higher in the former group, whereas the abundances of dendritic cells, B cells, and Treg cells in the spleen were significantly lower. Our results suggest that the gut microbiome influences arthritis susceptibility.
Figures




References
-
- Gregersen P. K., Silver J. & Winchester R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30, 1205–1213 (1987). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

