Secretion Chaperones PrsA2 and HtrA Are Required for Listeria monocytogenes Replication following Intracellular Induction of Virulence Factor Secretion
- PMID: 27481256
- PMCID: PMC5038075
- DOI: 10.1128/IAI.00312-16
Secretion Chaperones PrsA2 and HtrA Are Required for Listeria monocytogenes Replication following Intracellular Induction of Virulence Factor Secretion
Abstract
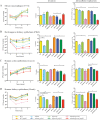
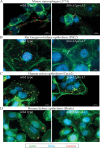
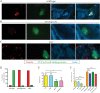
The Gram-positive bacterium Listeria monocytogenes transitions from an environmental organism to an intracellular pathogen following its ingestion by susceptible mammalian hosts. Bacterial replication within the cytosol of infected cells requires activation of the central virulence regulator PrfA followed by a PrfA-dependent induction of secreted virulence factors. The PrfA-induced secreted chaperone PrsA2 and the chaperone/protease HtrA contribute to the folding and stability of select proteins translocated across the bacterial membrane. L. monocytogenes strains that lack both prsA2 and htrA exhibit near-normal patterns of growth in broth culture but are severely attenuated in vivo We hypothesized that, in the absence of PrsA2 and HtrA, the increase in PrfA-dependent protein secretion that occurs following bacterial entry into the cytosol results in misfolded proteins accumulating at the bacterial membrane with a subsequent reduction in intracellular bacterial viability. Consistent with this hypothesis, the introduction of a constitutively activated allele of prfA (prfA*) into ΔprsA2 ΔhtrA strains was found to essentially inhibit bacterial growth at 37°C in broth culture. ΔprsA2 ΔhtrA strains were additionally found to be defective for cell invasion and vacuole escape in selected cell types, steps that precede full PrfA activation. These data establish the essential requirement for PrsA2 and HtrA in maintaining bacterial growth under conditions of PrfA activation. In addition, chaperone function is required for efficient bacterial invasion and rapid vacuole lysis within select host cell types, indicating roles for PrsA2/HtrA prior to cytosolic PrfA activation and the subsequent induction of virulence factor secretion.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Sauders BD, Durak MZ, Fortes E, Windham K, Schukken Y, Lembo AJ Jr, Akey B, Nightingale KK, Wiedmann M. 2006. Molecular characterization of Listeria monocytogenes from natural and urban environments. J Food Prot 69:93–105. - PubMed
-
- Centers for Disease Control and Prevention. 2011. Multistate outbreak of listeriosis associated with Jensen Farms cantaloupe–United States, August-September 2011. MMWR Morb Mortal Wkly Rep 60:1357–1358. - PubMed
-
- Olsen SJ, Patrick M, Hunter SB, Reddy V, Kornstein L, MacKenzie WR, Lane K, Bidol S, Stoltman GA, Frye DM, Lee I, Hurd S, Jones TF, LaPorte TN, Dewitt W, Graves L, Wiedmann M, Schoonmaker-Bopp DJ, Huang AJ, Vincent C, Bugenhagen A, Corby J, Carloni ER, Holcomb ME, Woron RF, Zansky SM, Dowdle G, Smith F, Ahrabi-Fard S, Ong AR, Tucker N, Hynes NA, Mead P. 2005. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin Infect Dis 40:962–967. doi:10.1086/428575. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

