Axitinib blocks Wnt/β-catenin signaling and directs asymmetric cell division in cancer
- PMID: 27482107
- PMCID: PMC4995957
- DOI: 10.1073/pnas.1604520113
Axitinib blocks Wnt/β-catenin signaling and directs asymmetric cell division in cancer
Abstract
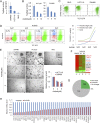
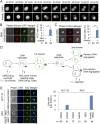
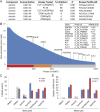
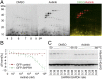
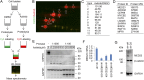
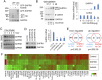
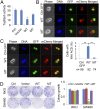
Oncogenic mutations of the Wnt (wingless)/β-catenin pathway are frequently observed in major cancer types. Thus far, however, no therapeutic agent targeting Wnt/β-catenin signaling is available for clinical use. Here we demonstrate that axitinib, a clinically approved drug, strikingly blocks Wnt/β-catenin signaling in cancer cells, zebrafish, and Apc(min/+) mice. Notably, axitinib dramatically induces Wnt asymmetry and nonrandom DNA segregation in cancer cells by promoting nuclear β-catenin degradation independent of the GSK3β (glycogen synthase kinase3β)/APC (adenomatous polyposis coli) complex. Using a DARTS (drug affinity-responsive target stability) assay coupled to 2D-DIGE (2D difference in gel electrophoresis) and mass spectrometry, we have identified the E3 ubiquitin ligase SHPRH (SNF2, histone-linker, PHD and RING finger domain-containing helicase) as the direct target of axitinib in blocking Wnt/β-catenin signaling. Treatment with axitinib stabilizes SHPRH and thereby increases the ubiquitination and degradation of β-catenin. Our findings suggest a previously unreported mechanism of nuclear β-catenin regulation and indicate that axitinib, a clinically approved drug, would provide therapeutic benefits for cancer patients with aberrant nuclear β-catenin activation.
Keywords: SHPRH; asymmetric cell division; axitinib; β-catenin.
Conflict of interest statement
The Technology Transfer Organization of University of Bergen (BTO) has filed a patent application (pending) for the use of axitinib as a WNT/β-catenin inhibitor in selected cancer types. None of the authors are affiliated with BTO. X.K., Y.Q., K.-H.K., and A.M.Ø are the inventors of the patent.
Figures







Similar articles
-
Regulation of Transcription Factor SP1 by the β-Catenin Destruction Complex Modulates Wnt Response.Mol Cell Biol. 2018 Oct 29;38(22):e00188-18. doi: 10.1128/MCB.00188-18. Print 2018 Nov 15. Mol Cell Biol. 2018. PMID: 30181396 Free PMC article.
-
E3 ubiquitin ligase Mule targets β-catenin under conditions of hyperactive Wnt signaling.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):E1148-E1157. doi: 10.1073/pnas.1621355114. Epub 2017 Jan 30. Proc Natl Acad Sci U S A. 2017. PMID: 28137882 Free PMC article.
-
The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of β-catenin.Mol Cell Biol. 2014 Dec 1;34(23):4355-66. doi: 10.1128/MCB.00731-14. Epub 2014 Sep 29. Mol Cell Biol. 2014. PMID: 25266658 Free PMC article.
-
Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer.Biomed Pharmacother. 2019 Feb;110:473-481. doi: 10.1016/j.biopha.2018.11.082. Epub 2018 Dec 6. Biomed Pharmacother. 2019. PMID: 30530050 Review.
-
New steps in the Wnt/beta-catenin signal transduction pathway.Recent Prog Horm Res. 2000;55:225-36. Recent Prog Horm Res. 2000. PMID: 11036939 Review.
Cited by
-
Antiangiogenic Compound Axitinib Demonstrates Low Toxicity and Antitumoral Effects against Medulloblastoma.Cancers (Basel). 2021 Dec 24;14(1):70. doi: 10.3390/cancers14010070. Cancers (Basel). 2021. PMID: 35008234 Free PMC article.
-
WNK regulates Wnt signalling and β-Catenin levels by interfering with the interaction between β-Catenin and GID.Commun Biol. 2020 Nov 12;3(1):666. doi: 10.1038/s42003-020-01386-2. Commun Biol. 2020. PMID: 33184430 Free PMC article.
-
The Zinc Finger Transcription Factor PLAGL2 Enhances Stem Cell Fate and Activates Expression of ASCL2 in Intestinal Epithelial Cells.Stem Cell Reports. 2018 Aug 14;11(2):410-424. doi: 10.1016/j.stemcr.2018.06.009. Epub 2018 Jul 12. Stem Cell Reports. 2018. PMID: 30017821 Free PMC article.
-
ALCAP2 inhibits lung adenocarcinoma cell proliferation, migration and invasion via the ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L.Cell Death Dis. 2021 Jul 31;12(8):755. doi: 10.1038/s41419-021-04043-6. Cell Death Dis. 2021. PMID: 34330894 Free PMC article.
-
MYB fusions and CD markers as tools for authentication and purification of cancer stem cells from salivary adenoid cystic carcinoma.Stem Cell Res. 2017 May;21:160-166. doi: 10.1016/j.scr.2017.05.002. Epub 2017 May 5. Stem Cell Res. 2017. PMID: 28500913 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

