Full-Length cDNA Cloning, Molecular Characterization and Differential Expression Analysis of Lysophospholipase I from Ovis aries
- PMID: 27483239
- PMCID: PMC5000604
- DOI: 10.3390/ijms17081206
Full-Length cDNA Cloning, Molecular Characterization and Differential Expression Analysis of Lysophospholipase I from Ovis aries
Abstract
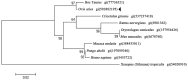
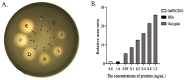
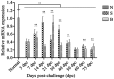
Lysophospholipase I (LYPLA1) is an important protein with multiple functions. In this study, the full-length cDNA of the LYPLA1 gene from Ovis aries (OaLypla1) was cloned using primers and rapid amplification of cDNA ends (RACE) technology. The full-length OaLypla1 was 2457 bp with a 5'-untranslated region (UTR) of 24 bp, a 3'-UTR of 1740 bp with a poly (A) tail, and an open reading frame (ORF) of 693 bp encoding a protein of 230 amino acid residues with a predicted molecular weight of 24,625.78 Da. Phylogenetic analysis showed that the OaLypla1 protein shared a high amino acid identity with LYPLA1 of Bos taurus. The recombinant OaLypla1 protein was expressed and purified, and its phospholipase activity was identified. Monoclonal antibodies (mAb) against OaLypla1 that bound native OaLypla1 were generated. Real-time PCR analysis revealed that OaLypla1 was constitutively expressed in the liver, spleen, lung, kidney, and white blood cells of sheep, with the highest level in the kidney. Additionally, the mRNA levels of OaLypla1 in the buffy coats of sheep challenged with virulent or avirulent Brucella strains were down-regulated compared to untreated sheep. The results suggest that OaLypla1 may have an important physiological role in the host response to bacteria. The function of OaLypla1 in the host response to bacterial infection requires further study in the future.
Keywords: Brucella; Lysophospholipase I; Ovis aries; differential expression; tissue distribution.
Figures





Similar articles
-
Full-length cDNA cloning, molecular characterization and differential expression analysis of peroxiredoxin 6 from Ovis aries.Vet Immunol Immunopathol. 2015 Apr 15;164(3-4):208-19. doi: 10.1016/j.vetimm.2015.01.006. Epub 2015 Jan 28. Vet Immunol Immunopathol. 2015. PMID: 25712755
-
Molecular characterization and differential expression analysis of interleukin 1β from Ovis aries.Microb Pathog. 2018 Mar;116:180-188. doi: 10.1016/j.micpath.2018.01.011. Epub 2018 Jan 10. Microb Pathog. 2018. PMID: 29331367
-
Molecular cloning, expression and characterization of programmed cell death 10 from sheep (Ovis aries).Gene. 2015 Mar 1;558(1):65-74. doi: 10.1016/j.gene.2014.12.040. Epub 2014 Dec 22. Gene. 2015. PMID: 25541025
-
Mammalian lysophospholipases.Biochim Biophys Acta. 1999 Jul 9;1439(1):1-16. doi: 10.1016/s1388-1981(99)00063-3. Biochim Biophys Acta. 1999. PMID: 10395961 Review. No abstract available.
-
An in Vivo Active Carbamate-based Dual Inhibitor of Lysophospholipase 1 (LYPLA1) and Lysophospholipase 2 (LYPLA2).2013 Dec 9 [updated 2014 Sep 18]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. 2013 Dec 9 [updated 2014 Sep 18]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. PMID: 25506974 Free Books & Documents. Review.
Cited by
-
A Novel, Rapid, and Simple PMA-qPCR Method for Detection and Counting of Viable Brucella Organisms.J Vet Res. 2020 May 12;64(2):253-261. doi: 10.2478/jvetres-2020-0033. eCollection 2020 Jun. J Vet Res. 2020. PMID: 32587912 Free PMC article.
References
-
- Portilla D., Crew M.D., Grant D., Serrero G., Bates L.M., Dai G., Sasner M., Cheng J., Buonanno A. cDNA cloning and expression of a novel family of enzymes with calcium-independent phospholipase A2 and lysophospholipase activities. J. Am. Soc. Nephrol. 1998;9:1178–1186. - PubMed
-
- Sugimoto H., Hayashi H., Yamashita S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 1996;271:7705–7711. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

