INC-Seq: accurate single molecule reads using nanopore sequencing
- PMID: 27485345
- PMCID: PMC4970289
- DOI: 10.1186/s13742-016-0140-7
INC-Seq: accurate single molecule reads using nanopore sequencing
Abstract
Background: Nanopore sequencing provides a rapid, cheap and portable real-time sequencing platform with the potential to revolutionize genomics. However, several applications are limited by relatively high single-read error rates (>10 %), including RNA-seq, haplotype sequencing and 16S sequencing.
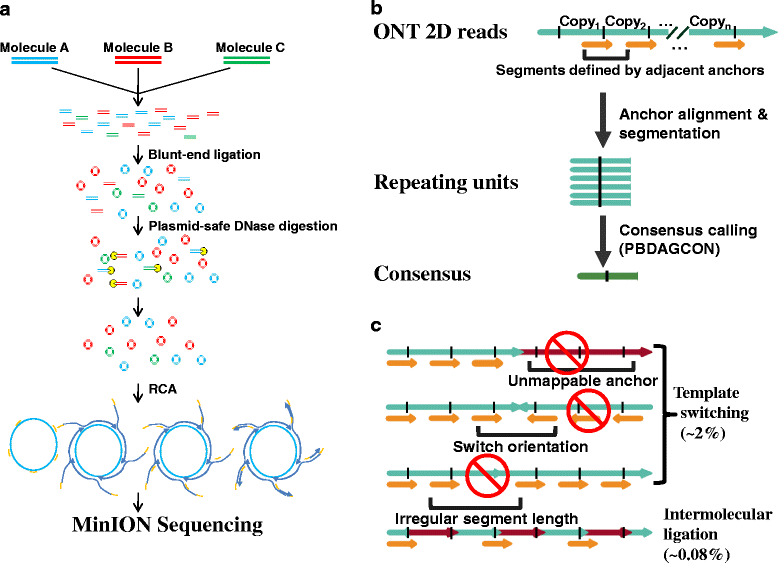
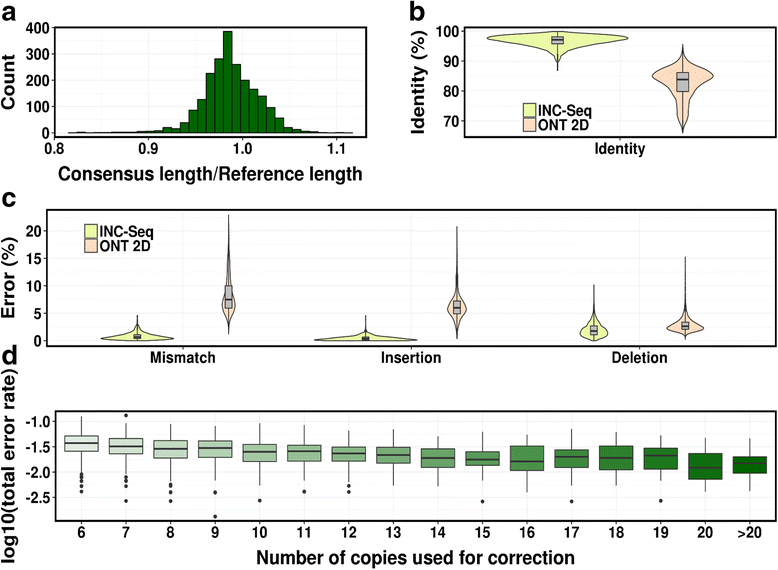
Results: We developed the Intramolecular-ligated Nanopore Consensus Sequencing (INC-Seq) as a strategy for obtaining long and accurate nanopore reads, starting with low input DNA. Applying INC-Seq for 16S rRNA-based bacterial profiling generated full-length amplicon sequences with a median accuracy >97 %.
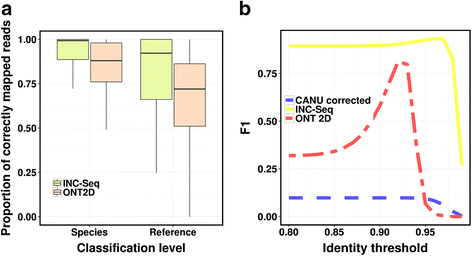
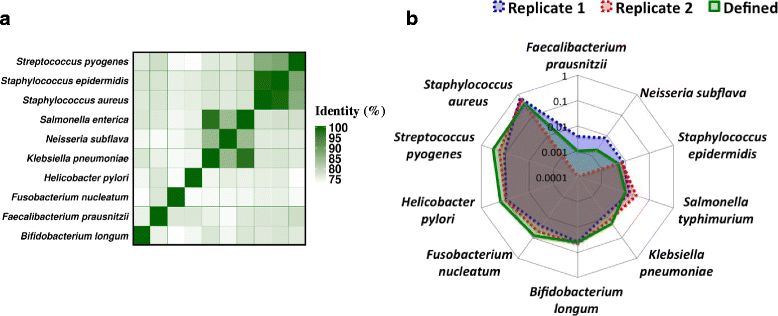
Conclusions: INC-Seq reads enabled accurate species-level classification, identification of species at 0.1 % abundance and robust quantification of relative abundances, providing a cheap and effective approach for pathogen detection and microbiome profiling on the MinION system.
Keywords: Barcode sequencing; Consensus algorithms; Nanopore sequencing; Rolling circle amplification.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

