Chemoinformatic expedition of the chemical space of fungal products
- PMID: 27485744
- PMCID: PMC5558535
- DOI: 10.4155/fmc-2016-0079
Chemoinformatic expedition of the chemical space of fungal products
Erratum in
-
Corrigendum.Future Med Chem. 2016 Nov;8(17):2167. doi: 10.4155/fmc-2016-0079c1. Future Med Chem. 2016. PMID: 27786551 Free PMC article. No abstract available.
Abstract
Aim: Fungi are valuable resources for bioactive secondary metabolites. However, the chemical space of fungal secondary metabolites has been studied only on a limited basis. Herein, we report a comprehensive chemoinformatic analysis of a unique set of 207 fungal metabolites isolated and characterized in a USA National Cancer Institute funded drug discovery project.
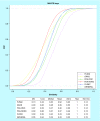
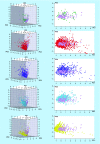
Results: Comparison of the molecular complexity of the 207 fungal metabolites with approved anticancer and nonanticancer drugs, compounds in clinical studies, general screening compounds and molecules Generally Recognized as Safe revealed that fungal metabolites have high degree of complexity. Molecular fingerprints showed that fungal metabolites are as structurally diverse as other natural products and have, in general, drug-like physicochemical properties.
Conclusion: Fungal products represent promising candidates to expand the medicinally relevant chemical space. This work is a significant expansion of an analysis reported years ago for a smaller set of compounds (less than half of the ones included in the present work) from filamentous fungi using different structural properties.
Keywords: chemical space; chemoinformatics; fungal metabolites; master key compound; molecular complexity; molecular fingerprint.
Conflict of interest statement
The authors thank the Universidad Nacional Autónoma de México (UNAM) for grant PAPIME No. PE200116, and the institutional program Nuevas Alternativas de Tratamiento para Enfermedades Infecciosas (NUATEI) of the Instituto de Investigaciones Biomédicas (IIB) UNAM for financial support, and the Consejo Nacional de Ciencia y Tecnología (CONACyT) for grant 236564. FDP-M is grateful to CONACyT for the fellowship 660465/576637. FD Prieto-Martínez is grateful to CONACyT for the fellowship No. 660465/576637. The isolation of fungal metabolites from the Mycosynthetix library via researchers at UNCG was funded by grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures




References
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. - PubMed
-
- Muller-Kuhrt L. Putting nature back into drug discovery. Nat. Biotech. 2003;21(6):602–602. - PubMed
-
- Strader CR, Pearce CJ, Oberlies NH. Fingolimod (FTY720): a recently approved multiple sclerosis drug based on a fungal secondary metabolite. J. Nat. Prod. 2011;74(4):900–907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
