Single-cell analyses of X Chromosome inactivation dynamics and pluripotency during differentiation
- PMID: 27486082
- PMCID: PMC5052059
- DOI: 10.1101/gr.201954.115
Single-cell analyses of X Chromosome inactivation dynamics and pluripotency during differentiation
Abstract
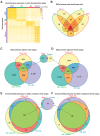
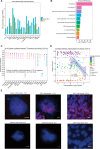
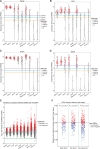
Pluripotency, differentiation, and X Chromosome inactivation (XCI) are key aspects of embryonic development. However, the underlying relationship and mechanisms among these processes remain unclear. Here, we systematically dissected these features along developmental progression using mouse embryonic stem cells (mESCs) and single-cell RNA sequencing with allelic resolution. We found that mESCs grown in a ground state 2i condition displayed transcriptomic profiles diffused from preimplantation mouse embryonic cells, whereas EpiStem cells closely resembled the post-implantation epiblast. Sex-related gene expression varied greatly across distinct developmental states. We also identified novel markers that were highly enriched in each developmental state. Moreover, we revealed that several novel pathways, including PluriNetWork and Focal Adhesion, were responsible for the delayed progression of female EpiStem cells. Importantly, we "digitalized" XCI progression using allelic expression of active and inactive X Chromosomes and surprisingly found that XCI states exhibited profound variability in each developmental state, including the 2i condition. XCI progression was not tightly synchronized with loss of pluripotency and increase of differentiation at the single-cell level, although these processes were globally correlated. In addition, highly expressed genes, including core pluripotency factors, were in general biallelically expressed. Taken together, our study sheds light on the dynamics of XCI progression and the asynchronicity between pluripotency, differentiation, and XCI.
© 2016 Chen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Bradley A, Evans M, Kaufman MH, Robertson E. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309: 255–256. - PubMed
-
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun BW, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
