Effect of Shear Stress on Pseudomonas aeruginosa Isolated from the Cystic Fibrosis Lung
- PMID: 27486191
- PMCID: PMC4981712
- DOI: 10.1128/mBio.00813-16
Effect of Shear Stress on Pseudomonas aeruginosa Isolated from the Cystic Fibrosis Lung
Abstract
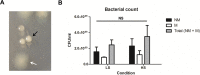
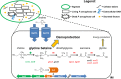
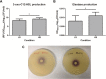
Chronic colonization of the lungs by Pseudomonas aeruginosa is one of the major causes of morbidity and mortality in cystic fibrosis (CF) patients. To gain insights into the characteristic biofilm phenotype of P. aeruginosa in the CF lungs, mimicking the CF lung environment is critical. We previously showed that growth of the non-CF-adapted P. aeruginosa PAO1 strain in a rotating wall vessel, a device that simulates the low fluid shear (LS) conditions present in the CF lung, leads to the formation of in-suspension, self-aggregating biofilms. In the present study, we determined the phenotypic and transcriptomic changes associated with the growth of a highly adapted, transmissible P. aeruginosa CF strain in artificial sputum medium under LS conditions. Robust self-aggregating biofilms were observed only under LS conditions. Growth under LS conditions resulted in the upregulation of genes involved in stress response, alginate biosynthesis, denitrification, glycine betaine biosynthesis, glycerol metabolism, and cell shape maintenance, while genes involved in phenazine biosynthesis, type VI secretion, and multidrug efflux were downregulated. In addition, a number of small RNAs appeared to be involved in the response to shear stress. Finally, quorum sensing was found to be slightly but significantly affected by shear stress, resulting in higher production of autoinducer molecules during growth under high fluid shear (HS) conditions. In summary, our study revealed a way to modulate the behavior of a highly adapted P. aeruginosa CF strain by means of introducing shear stress, driving it from a biofilm lifestyle to a more planktonic lifestyle.
Importance: Biofilm formation by Pseudomonas aeruginosa is one of the hallmarks of chronic cystic fibrosis (CF) lung infections. The biofilm matrix protects this bacterium from antibiotics as well as from the immune system. Hence, the prevention or reversion of biofilm formation is believed to have a great impact on treatment of chronic P. aeruginosa CF lung infections. In the present study, we showed that it is possible to modulate the behavior of a highly adapted transmissible P. aeruginosa CF isolate at both the transcriptomic and phenotypic levels by introducing shear stress in a CF-like environment, driving it from a biofilm to a planktonic lifestyle. Consequently, the results obtained in this study are of great importance with regard to therapeutic applications that introduce shear stress in the lungs of CF patients.
Copyright © 2016 Dingemans et al.
Figures


Similar articles
-
Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01.Environ Microbiol. 2008 Aug;10(8):2098-110. doi: 10.1111/j.1462-2920.2008.01631.x. Epub 2008 Apr 22. Environ Microbiol. 2008. PMID: 18430020
-
Unveiling the early events of Pseudomonas aeruginosa adaptation in cystic fibrosis airway environment using a long-term in vitro maintenance.Int J Med Microbiol. 2018 Dec;308(8):1053-1064. doi: 10.1016/j.ijmm.2018.10.003. Epub 2018 Oct 11. Int J Med Microbiol. 2018. PMID: 30377031
-
Glycerol metabolism promotes biofilm formation by Pseudomonas aeruginosa.Can J Microbiol. 2016 Aug;62(8):704-10. doi: 10.1139/cjm-2016-0119. Epub 2016 May 3. Can J Microbiol. 2016. PMID: 27392247
-
Airway biofilms: implications for pathogenesis and therapy of respiratory tract infections.Treat Respir Med. 2005;4(4):241-53. doi: 10.2165/00151829-200504040-00003. Treat Respir Med. 2005. PMID: 16086598 Review.
-
Pseudomonas aeruginosa biofilms in cystic fibrosis.Future Microbiol. 2010 Nov;5(11):1663-74. doi: 10.2217/fmb.10.125. Future Microbiol. 2010. PMID: 21133688 Review.
Cited by
-
Biofloc-Based Enhanced Survival of Litopenaeus vannamei Upon AHPND-Causing Vibrio parahaemolyticus Challenge Is Partially Mediated by Reduced Expression of Its Virulence Genes.Front Microbiol. 2020 Jun 24;11:1270. doi: 10.3389/fmicb.2020.01270. eCollection 2020. Front Microbiol. 2020. PMID: 32670225 Free PMC article.
-
Physical communication pathways in bacteria: an extra layer to quorum sensing.Biophys Rev. 2025 Mar 4;17(2):667-685. doi: 10.1007/s12551-025-01290-1. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376406 Free PMC article. Review.
-
Calling all hosts: Bacterial communication in situ.Chem. 2017 Mar 9;2(3):334-358. doi: 10.1016/j.chempr.2017.02.001. Chem. 2017. PMID: 28948238 Free PMC article.
-
Transcriptional profiling of the mutualistic bacterium Vibrio fischeri and an hfq mutant under modeled microgravity.NPJ Microgravity. 2018 Dec 18;4:25. doi: 10.1038/s41526-018-0060-1. eCollection 2018. NPJ Microgravity. 2018. PMID: 30588486 Free PMC article.
-
Pseudomonas aeruginosa in the Cystic Fibrosis Lung.Adv Exp Med Biol. 2022;1386:347-369. doi: 10.1007/978-3-031-08491-1_13. Adv Exp Med Biol. 2022. PMID: 36258079
References
-
- Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105:3100–3105. doi:10.1073/pnas.0711982105. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

