Comprehensive Identification of Meningococcal Genes and Small Noncoding RNAs Required for Host Cell Colonization
- PMID: 27486197
- PMCID: PMC4981724
- DOI: 10.1128/mBio.01173-16
Comprehensive Identification of Meningococcal Genes and Small Noncoding RNAs Required for Host Cell Colonization
Abstract
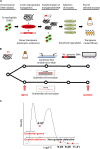
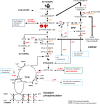
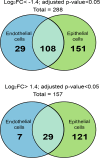
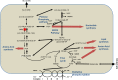
Neisseria meningitidis is a leading cause of bacterial meningitis and septicemia, affecting infants and adults worldwide. N. meningitidis is also a common inhabitant of the human nasopharynx and, as such, is highly adapted to its niche. During bacteremia, N. meningitidis gains access to the blood compartment, where it adheres to endothelial cells of blood vessels and causes dramatic vascular damage. Colonization of the nasopharyngeal niche and communication with the different human cell types is a major issue of the N. meningitidis life cycle that is poorly understood. Here, highly saturated random transposon insertion libraries of N. meningitidis were engineered, and the fitness of mutations during routine growth and that of colonization of endothelial and epithelial cells in a flow device were assessed in a transposon insertion site sequencing (Tn-seq) analysis. This allowed the identification of genes essential for bacterial growth and genes specifically required for host cell colonization. In addition, after having identified the small noncoding RNAs (sRNAs) located in intergenic regions, the phenotypes associated with mutations in those sRNAs were defined. A total of 383 genes and 8 intergenic regions containing sRNA candidates were identified to be essential for growth, while 288 genes and 33 intergenic regions containing sRNA candidates were found to be specifically required for host cell colonization.
Importance: Meningococcal meningitis is a common cause of meningitis in infants and adults. Neisseria meningitidis (meningococcus) is also a commensal bacterium of the nasopharynx and is carried by 3 to 30% of healthy humans. Under some unknown circumstances, N. meningitidis is able to invade the bloodstream and cause either meningitis or a fatal septicemia known as purpura fulminans. The onset of symptoms is sudden, and death can follow within hours. Although many meningococcal virulence factors have been identified, the mechanisms that allow the bacterium to switch from the commensal to pathogen state remain unknown. Therefore, we used a Tn-seq strategy coupled to high-throughput DNA sequencing technologies to find genes for proteins used by N. meningitidis to specifically colonize epithelial cells and primary brain endothelial cells. We identified 383 genes and 8 intergenic regions containing sRNAs essential for growth and 288 genes and 33 intergenic regions containing sRNAs required specifically for host cell colonization.
Copyright © 2016 Capel et al.
Figures

Similar articles
-
Identification of genes involved in Neisseria meningitidis colonization.Infect Immun. 2013 Sep;81(9):3375-81. doi: 10.1128/IAI.00421-13. Epub 2013 Jul 1. Infect Immun. 2013. PMID: 23817612 Free PMC article.
-
The Meningococcal Cysteine Transport System Plays a Crucial Role in Neisseria meningitidis Survival in Human Brain Microvascular Endothelial Cells.mBio. 2018 Dec 11;9(6):e02332-18. doi: 10.1128/mBio.02332-18. mBio. 2018. PMID: 30538184 Free PMC article.
-
Neisseria meningitidis Sibling Small Regulatory RNAs Connect Metabolism with Colonization by Controlling Propionate Use.J Bacteriol. 2023 Mar 21;205(3):e0046222. doi: 10.1128/jb.00462-22. Epub 2023 Mar 1. J Bacteriol. 2023. PMID: 36856428 Free PMC article.
-
An Overview of Neisseria meningitidis.Methods Mol Biol. 2019;1969:1-16. doi: 10.1007/978-1-4939-9202-7_1. Methods Mol Biol. 2019. PMID: 30877666 Review.
-
Molecular interactions between Neisseria meningitidis and its human host.Cell Microbiol. 2019 Nov;21(11):e13063. doi: 10.1111/cmi.13063. Epub 2019 Jun 13. Cell Microbiol. 2019. PMID: 31167044 Free PMC article. Review.
Cited by
-
A Comprehensive Overview of Online Resources to Identify and Predict Bacterial Essential Genes.Front Microbiol. 2017 Nov 27;8:2331. doi: 10.3389/fmicb.2017.02331. eCollection 2017. Front Microbiol. 2017. PMID: 29230204 Free PMC article. Review.
-
Sinorhizobium meliloti BR-bodies promote fitness during host colonization.bioRxiv [Preprint]. 2024 Apr 6:2024.04.05.588320. doi: 10.1101/2024.04.05.588320. bioRxiv. 2024. PMID: 38617242 Free PMC article. Preprint.
-
Peripheral blood vessels are a niche for blood-borne meningococci.Virulence. 2017 Nov 17;8(8):1808-1819. doi: 10.1080/21505594.2017.1391446. Epub 2017 Nov 27. Virulence. 2017. PMID: 29099305 Free PMC article.
-
Establishing the critical role of peripheral blood vessel colonisation by Neisseria meningitidis in invasive meningococcal disease.Virulence. 2017 Nov 17;8(8):1513-1515. doi: 10.1080/21505594.2017.1415191. Virulence. 2017. PMID: 29219762 Free PMC article.
-
A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination.PLoS Comput Biol. 2018 Feb 5;14(2):e1005958. doi: 10.1371/journal.pcbi.1005958. eCollection 2018 Feb. PLoS Comput Biol. 2018. PMID: 29401456 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

