Phosphoproteomics to Characterize Host Response During Influenza A Virus Infection of Human Macrophages
- PMID: 27486199
- PMCID: PMC5054344
- DOI: 10.1074/mcp.M116.057984
Phosphoproteomics to Characterize Host Response During Influenza A Virus Infection of Human Macrophages
Abstract
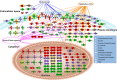
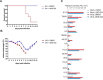
Influenza A viruses cause infections in the human respiratory tract and give rise to annual seasonal outbreaks, as well as more rarely dreaded pandemics. Influenza A viruses become quickly resistant to the virus-directed antiviral treatments, which are the current main treatment options. A promising alternative approach is to target host cell factors that are exploited by influenza viruses. To this end, we characterized the phosphoproteome of influenza A virus infected primary human macrophages to elucidate the intracellular signaling pathways and critical host factors activated upon influenza infection. We identified 1675 phosphoproteins, 4004 phosphopeptides and 4146 nonredundant phosphosites. The phosphorylation of 1113 proteins (66%) was regulated upon infection, highlighting the importance of such global phosphoproteomic profiling in primary cells. Notably, 285 of the identified phosphorylation sites have not been previously described in publicly available phosphorylation databases, despite many published large-scale phosphoproteome studies using human and mouse cell lines. Systematic bioinformatics analysis of the phosphoproteome data indicated that the phosphorylation of proteins involved in the ubiquitin/proteasome pathway (such as TRIM22 and TRIM25) and antiviral responses (such as MAVS) changed in infected macrophages. Proteins known to play roles in small GTPase-, mitogen-activated protein kinase-, and cyclin-dependent kinase- signaling were also regulated by phosphorylation upon infection. In particular, the influenza infection had a major influence on the phosphorylation profiles of a large number of cyclin-dependent kinase substrates. Functional studies using cyclin-dependent kinase inhibitors showed that the cyclin-dependent kinase activity is required for efficient viral replication and for activation of the host antiviral responses. In addition, we show that cyclin-dependent kinase inhibitors protect IAV-infected mice from death. In conclusion, we provide the first comprehensive phosphoproteome characterization of influenza A virus infection in primary human macrophages, and provide evidence that cyclin-dependent kinases represent potential therapeutic targets for more effective treatment of influenza infections.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Phosphoproteome Analysis of Cells Infected with Adapted and Nonadapted Influenza A Virus Reveals Novel Pro- and Antiviral Signaling Networks.J Virol. 2019 Jun 14;93(13):e00528-19. doi: 10.1128/JVI.00528-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996098 Free PMC article.
-
Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages.PLoS Pathog. 2011 May;7(5):e1001340. doi: 10.1371/journal.ppat.1001340. Epub 2011 May 12. PLoS Pathog. 2011. PMID: 21589892 Free PMC article.
-
Phosphoproteomic-based kinase profiling early in influenza virus infection identifies GRK2 as antiviral drug target.Nat Commun. 2018 Sep 11;9(1):3679. doi: 10.1038/s41467-018-06119-y. Nat Commun. 2018. PMID: 30206219 Free PMC article.
-
Influenza Virus Infections and Cellular Kinases.Viruses. 2019 Feb 20;11(2):171. doi: 10.3390/v11020171. Viruses. 2019. PMID: 30791550 Free PMC article. Review.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
Cited by
-
Multi-Omics Studies towards Novel Modulators of Influenza A Virus-Host Interaction.Viruses. 2016 Sep 29;8(10):269. doi: 10.3390/v8100269. Viruses. 2016. PMID: 27690086 Free PMC article. Review.
-
TMT-based quantitative proteomics analysis reveals the attenuated replication mechanism of Newcastle disease virus caused by nuclear localization signal mutation in viral matrix protein.Virulence. 2020 Dec;11(1):607-635. doi: 10.1080/21505594.2020.1770482. Virulence. 2020. PMID: 32420802 Free PMC article.
-
Host Gene Expression of Macrophages in Response to Feline Coronavirus Infection.Cells. 2020 Jun 9;9(6):1431. doi: 10.3390/cells9061431. Cells. 2020. PMID: 32526950 Free PMC article.
-
Neoadjuvant chemotherapy is associated with an altered metabolic profile and increased cancer stemness in patients with pancreatic ductal adenocarcinoma.Mol Oncol. 2023 Jan;17(1):59-81. doi: 10.1002/1878-0261.13344. Epub 2022 Dec 5. Mol Oncol. 2023. PMID: 36400567 Free PMC article.
-
Multi-omics data integration reveals the complexity and diversity of host factors associated with influenza virus infection.PeerJ. 2023 Oct 9;11:e16194. doi: 10.7717/peerj.16194. eCollection 2023. PeerJ. 2023. PMID: 37842064 Free PMC article.
References
-
- Flannery B., Clippard J., Zimmerman R. K., Nowalk M. P., Jackson M. L., Jackson L. A., Monto A. S., Petrie J. G., McLean H. Q., Belongia E. A., Gaglani M., Berman L., Foust A., Sessions W., Thaker S. N., Spencer S., Fry A. M., and Centers for Disease Control and Prevention.(2015) Early estimates of seasonal influenza vaccine effectiveness - united states, january 2015. MMWR Morb. Mortal. Wkly. Rep. 64, 10–15 - PMC - PubMed
-
- Spanakis N., Pitiriga V., Gennimata V., and Tsakris A. (2014) A review of neuraminidase inhibitor susceptibility in influenza strains. Expert Rev. Anti Infect. Ther. 12, 1325–1336 - PubMed
-
- Hurt A. C. (2014) The epidemiology and spread of drug resistant human influenza viruses. Curr. Opin. Virol. 8C, 22–29 - PubMed
-
- Meissner F., and Mann M. (2014) Quantitative shotgun proteomics: Considerations for a high-quality workflow in immunology. Nat. Immunol. 15, 112–117 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

