Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks
- PMID: 27486230
- PMCID: PMC5004913
- DOI: 10.1242/dev.138263
Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks
Abstract
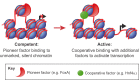
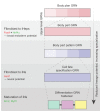
Although many approaches have been employed to generate defined fate in vitro, the resultant cells often appear developmentally immature or incompletely specified, limiting their utility. Growing evidence suggests that current methods of direct lineage conversion may rely on the transition through a developmental intermediate. Here, I hypothesize that complete conversion between cell fates is more probable and feasible via reversion to a developmentally immature state. I posit that this is due to the role of pioneer transcription factors in engaging silent, unmarked chromatin and activating hierarchical gene regulatory networks responsible for embryonic patterning. Understanding these developmental contexts will be essential for the precise engineering of cell identity.
Keywords: Cell fate engineering; Direct lineage reprogramming; Gene regulatory networks; Pioneer transcription factors.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The author declares no competing or financial interests.
Figures


References
-
- Bar-Nur O., Verheul C., Sommer A. G., Brumbaugh J., Schwarz B. A., Lipchina I., Huebner A. J., Mostoslavsky G. and Hochedlinger K. (2015). Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 33, 761-768. 10.1038/nbt.3247 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

