A Novel Role for Banana MaASR in the Regulation of Flowering Time in Transgenic Arabidopsis
- PMID: 27486844
- PMCID: PMC4972433
- DOI: 10.1371/journal.pone.0160690
A Novel Role for Banana MaASR in the Regulation of Flowering Time in Transgenic Arabidopsis
Abstract
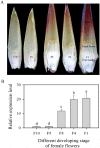
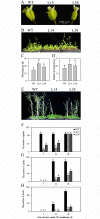
The abscisic acid (ABA)-, stress-, and ripening-induced (ASR) protein is a plant-specific hydrophilic transcriptional factor involved in fruit ripening and the abiotic stress response. To date, there have been no studies on the role of ASR genes in delayed flowering time. Here, we found that the ASR from banana, designated as MaASR, was preferentially expressed in the banana female flowers from the eighth, fourth, and first cluster of the inflorescence. MaASR transgenic lines (L14 and L38) had a clear delayed-flowering phenotype. The number of rosette leaves, sepals, and pedicel trichomes in L14 and L38 was greater than in the wild type (WT) under long day (LD) conditions. The period of buds, mid-flowers, and full bloom of L14 and L38 appeared later than the WT. cDNA microarray and quantitative real-time PCR (qRT-PCR) analyses revealed that overexpression of MaASR delays flowering through reduced expression of several genes, including photoperiod pathway genes, vernalization pathway genes, gibberellic acid pathway genes, and floral integrator genes, under short days (SD) for 28 d (from vegetative to reproductive transition stage); however, the expression of the autonomous pathway genes was not affected. This study provides the first evidence of a role for ASR genes in delayed flowering time in plants.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

