Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features
- PMID: 27486981
- PMCID: PMC5302970
- DOI: 10.18632/oncotarget.10937
Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features
Abstract
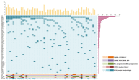
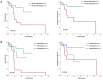
Clonal cytogenetic abnormalities are found in 20-30% of patients with chronic myelomonocytic leukemia (CMML), while gene mutations are present in >90% of cases. Patients with low risk cytogenetic features account for 80% of CMML cases and often fall into the low risk categories of CMML prognostic scoring systems, but the outcome differs considerably among them. We performed targeted deep sequencing of 83 myeloid-related genes in 56 CMML patients with low risk cytogenetic features or uninformative conventional cytogenetics (CC) at diagnosis, with the aim to identify the genetic characteristics of patients with a more aggressive disease. Targeted sequencing was also performed in a subset of these patients at time of acute myeloid leukemia (AML) transformation. Overall, 98% of patients harbored at least one mutation. Mutations in cell signaling genes were acquired at time of AML progression. Mutations in ASXL1, EZH2 and NRAS correlated with higher risk features and shorter overall survival (OS) and progression free survival (PFS). Patients with SRSF2 mutations associated with poorer OS, while absence of TET2 mutations (TET2wt) was predictive of shorter PFS. A decrease in OS and PFS was observed as the number of adverse risk gene mutations (ASXL1, EZH2, NRAS and SRSF2) increased. On multivariate analyses, CMML-specific scoring system (CPSS) and presence of adverse risk gene mutations remained significant for OS, while CPSS and TET2wt were predictive of PFS. These results confirm that mutation analysis can add prognostic value to patients with CMML and low risk cytogenetic features or uninformative CC.
Keywords: chronic myelomonocytic leukemia; gene mutations; normal karyotype; prognostic factors; targeted deep sequencing.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures
References
-
- Orazi A, Bennett JM, Germing U, Brunning RD, Bain BJ, Thiele J. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC; 2008. Chronic Myelomonocytic Leukemia.
-
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick H, Sultan C, Cox C. The chronic myeloid leukaemias: guidelines for distinguishing chronic granulocytic, atypical chronic myeloid, and chronic myelomonocytic leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br J Haematol. 1994;87:746–54. - PubMed
-
- Schuler E, Schroeder M, Neukirchen J, Strupp C, Xicoy B, Kündgen A, Hildebrandt B, Haas R, Gattermann N, Germing U. Refined medullary blast and white blood cell count based classification of chronic myelomonocytic leukemias. Leuk Res. 2014;38:1413–9. doi: 10.1016/j.leukres.2014.09.003. - DOI - PubMed
-
- Such E, Cervera J, Costa D, Solé F, Vallespí T, Luño E, Collado R, Calasanz MJ, Hernández-Rivas JM, Cigudosa JC, Nomdedeu B, Mallo M, Carbonell F, et al. Cytogenetic risk stratification in chronic myelomonocytic leukemia. Haematologica. 2011;96:375–83. doi: 10.3324/haematol.2010.030957. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

