Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas
- PMID: 27486988
- PMCID: PMC5312306
- DOI: 10.18632/oncotarget.10962
Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas
Abstract
Background: We previously identified PRAME as a biomarker for metastatic risk in Class 1 uveal melanomas. In this study, we sought to define a threshold value for positive PRAME expression (PRAME+) in a large dataset, identify factors associated with PRAME expression, evaluate the prognostic value of PRAME in Class 2 uveal melanomas, and determine whether PRAME expression is associated with aberrant hypomethylation of the PRAME promoter.
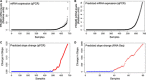
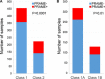
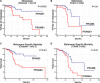
Results: Among 678 samples analyzed by qPCR, 498 (73.5%) were PRAME- and 180 (26.5%) were PRAME+. Class 1 tumors were more likely to be PRAME-, whereas Class 2 tumors were more likely to be PRAME+ (P < 0.0001). PRAME expression was associated with shorter time to metastasis and melanoma specific mortality in Class 2 tumors (P = 0.01 and P = 0.02, respectively). In Class 1 tumors, PRAME expression was directly associated with SF3B1 mutations (P < 0.0001) and inversely associated with EIF1AX mutations (P = 0.004). PRAME expression was strongly associated with hypomethylation at 12 CpG sites near the PRAME promoter.
Materials and methods: Analyses included PRAME mRNA expression, Class 1 versus Class 2 status, chromosomal copy number, mutation status of BAP1, EIF1AX, GNA11, GNAQ and SF3B1, and genomic DNA methylation status. Analyses were performed on 555 de-identified samples from Castle Biosciences, 123 samples from our center, and 80 samples from the TCGA.
Conclusions: PRAME is aberrantly hypomethylated and activated in Class 1 and Class 2 uveal melanomas and is associated with increased metastatic risk in both classes. Since PRAME has been successfully targeted for immunotherapy, it may prove to be a companion prognostic biomarker.
Keywords: DNA methylation; PRAME; chromosomal instability; preferentially expressed antigen in melanoma; uveal melanoma.
Conflict of interest statement
Dr. Harbour is the inventor of intellectual property related to the gene expression profile technology used in the study. Drs. Harbour and Bowcock are the inventors of intellectual property related to the discovery of
Figures


References
-
- Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, Aaberg TM, Jr, Altaweel MM, Bardenstein DS, Finger PT, Gallie BL, Harocopos GJ, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–1603. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

