Cotranslational signal-independent SRP preloading during membrane targeting
- PMID: 27487213
- PMCID: PMC5120976
- DOI: 10.1038/nature19309
Cotranslational signal-independent SRP preloading during membrane targeting
Abstract
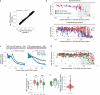
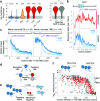
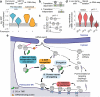
Ribosome-associated factors must properly decode the limited information available in nascent polypeptides to direct them to their correct cellular fate. It is unclear how the low complexity information exposed by the nascent chain suffices for accurate recognition by the many factors competing for the limited surface near the ribosomal exit site. Questions remain even for the well-studied cotranslational targeting cycle to the endoplasmic reticulum, involving recognition of linear hydrophobic signal sequences or transmembrane domains by the signal recognition particle (SRP). Notably, the SRP has low abundance relative to the large number of ribosome-nascent-chain complexes (RNCs), yet it accurately selects those destined for the endoplasmic reticulum. Despite their overlapping specificities, the SRP and the cotranslationally acting Hsp70 display precise mutually exclusive selectivity in vivo for their cognate RNCs. To understand cotranslational nascent chain recognition in vivo, here we investigate the cotranslational membrane-targeting cycle using ribosome profiling in yeast cells coupled with biochemical fractionation of ribosome populations. We show that the SRP preferentially binds secretory RNCs before their targeting signals are translated. Non-coding mRNA elements can promote this signal-independent pre-recruitment of SRP. Our study defines the complex kinetic interaction between elongation in the cytosol and determinants in the polypeptide and mRNA that modulate SRP–substrate selection and membrane targeting.
Figures








References
-
- Bornemann T, Holtkamp W, Wintermeyer W. Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat Commun. 2014;5:4180. - PubMed
Methods Online References
-
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. - PubMed
-
- Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. - PubMed
-
- Zhong T, Arndt KT. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell. 1993;73:1175–1186. - PubMed
-
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. - PubMed
-
- Freidlin PJ, Patterson RJ. Heparin releases monosomes and polysomes from rough endoplasmic reticulum. Biochem Biophys Res Commun. 1980;93:521–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

