Metrics for the Human Proteome Project 2016: Progress on Identifying and Characterizing the Human Proteome, Including Post-Translational Modifications
- PMID: 27487407
- PMCID: PMC5129622
- DOI: 10.1021/acs.jproteome.6b00511
Metrics for the Human Proteome Project 2016: Progress on Identifying and Characterizing the Human Proteome, Including Post-Translational Modifications
Abstract
The HUPO Human Proteome Project (HPP) has two overall goals: (1) stepwise completion of the protein parts list-the draft human proteome including confidently identifying and characterizing at least one protein product from each protein-coding gene, with increasing emphasis on sequence variants, post-translational modifications (PTMs), and splice isoforms of those proteins; and (2) making proteomics an integrated counterpart to genomics throughout the biomedical and life sciences community. PeptideAtlas and GPMDB reanalyze all major human mass spectrometry data sets available through ProteomeXchange with standardized protocols and stringent quality filters; neXtProt curates and integrates mass spectrometry and other findings to present the most up to date authorative compendium of the human proteome. The HPP Guidelines for Mass Spectrometry Data Interpretation version 2.1 were applied to manuscripts submitted for this 2016 C-HPP-led special issue [ www.thehpp.org/guidelines ]. The Human Proteome presented as neXtProt version 2016-02 has 16,518 confident protein identifications (Protein Existence [PE] Level 1), up from 13,664 at 2012-12, 15,646 at 2013-09, and 16,491 at 2014-10. There are 485 proteins that would have been PE1 under the Guidelines v1.0 from 2012 but now have insufficient evidence due to the agreed-upon more stringent Guidelines v2.0 to reduce false positives. neXtProt and PeptideAtlas now both require two non-nested, uniquely mapping (proteotypic) peptides of at least 9 aa in length. There are 2,949 missing proteins (PE2+3+4) as the baseline for submissions for this fourth annual C-HPP special issue of Journal of Proteome Research. PeptideAtlas has 14,629 canonical (plus 1187 uncertain and 1755 redundant) entries. GPMDB has 16,190 EC4 entries, and the Human Protein Atlas has 10,475 entries with supportive evidence. neXtProt, PeptideAtlas, and GPMDB are rich resources of information about post-translational modifications (PTMs), single amino acid variants (SAAVSs), and splice isoforms. Meanwhile, the Biology- and Disease-driven (B/D)-HPP has created comprehensive SRM resources, generated popular protein lists to guide targeted proteomics assays for specific diseases, and launched an Early Career Researchers initiative.
Keywords: GPMDB; Human Protein Atlas; N-termini; PTMs (post-translational modifications); PeptideAtlas; SAAV (single amino acid variants); guidelines; metrics; neXtProt; splice isoforms.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
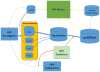
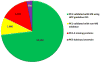
References
-
- Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS. The Human Proteome Project: current state and future direction. Mol Cell Proteomics. 2011;10(7):M111 009993. - PMC - PubMed
-
- Marko-Varga G, Omenn GS, Paik YK, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res. 2013;12(1):1–5. - PubMed
-
- Deutsch EW, Overall CM, Van Eyk J, Baker M, Paik YK, Weintraub S, Lane L, Martens L, Vandenbrouck Y, Kusebauch U, Hancock W, Hermjakob H, Aebersold R, Moritz RL, Omenn GS. Human Proteome Project mass spectrometry data interpretation guidelines 2.1. J Proteome Res. 2016 doi: 10.1021/acs.jproteo-me.6b00392. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

